The Drosophila histone variant H2Av facilitates Notch signaling activity in a two-tier regulatory fashion
- PMID: 40598498
- PMCID: PMC12220669
- DOI: 10.1186/s12964-025-02333-6
The Drosophila histone variant H2Av facilitates Notch signaling activity in a two-tier regulatory fashion
Abstract
Background: H2Av is an evolutionarily conserved H2A variant protein involved in the regulation of transcription. The Tip60 complex is recruited by different transcription factors to facilitate the incorporation and acetylation of H2Av, thereby influencing target gene expression. The Tip60-H2Av axis is involved in various developmental processes, though its precise roles are not yet fully understood.
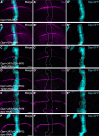
Methods: RNA interference and gene mutation technology were used to screen essential genes in regulating Notch signaling pathway. Immunostaining was used to detect the protein level of H2Av, Tip60 complex as well as Notch signaling pathway components. Chromatin immunoprecipitation assays were performed to detect the specific binding site of H2Av in E(spl)-Complex and Su(H) genes.
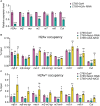
Result: Here we report that H2Av is required for Notch signaling activation during Drosophila wing development. H2Av depletion disrupts the expression of Notch target genes, resulting in wing marginal defects. Unexpectedly, we find that H2Av regulates the expression of the Su(H) gene which encodes for the transcription factor of the Notch signaling cascade. We further demonstrate that the Tip60 complex modulates the transcription of both Notch targets and Su(H) likely through H2Av. Based on these observations, we propose a model that the Tip60-H2Av axis facilitates Notch pathway activation by simultaneously promoting the expression of both the target genes and the transcription factor.
Conclusion: This study offers insights into the diverse roles of the Tip60-H2Av axis in Notch pathway activation by identifying a novel two-tier regulatory mechanism which may also be utilized by other chromatin remodeling factors.
Keywords: Drosophila; H2Av; Notch; Su(H); Tip60.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures






References
-
- Millán-Zambrano G, Burton A, Bannister AJ, Schneider R. Histone post-translational modifications - cause and consequence of genome function. Nat Rev Genet. 2022;23:563–80. - PubMed
-
- Li M, Fang Y. Histone variants: the artists of eukaryotic chromatin. Sci China Life Sci. 2015;58:232–9. - PubMed
-
- Biterge B, Schneider R. Histone variants: key players of chromatin. Cell Tissue Res. 2014;356:457–66. - PubMed
-
- Talbert PB, Henikoff S. Histone variants–ancient wrap artists of the epigenome. Nat Rev Mol Cell Biol. 2010;11:264–75. - PubMed
-
- Zlatanova J, Thakar A. H2A.Z: view from the top. Structure. 2008;16:166–79. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

