A-to-I mRNA editing in bacteria can affect protein sequence, disulfide bond formation, and function
- PMID: 40598894
- PMCID: PMC12214031
- DOI: 10.1093/nar/gkaf584
A-to-I mRNA editing in bacteria can affect protein sequence, disulfide bond formation, and function
Abstract
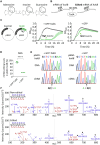
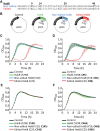
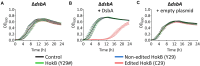
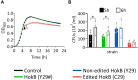
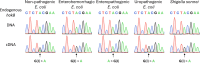
Adenosine-to-inosine (A-to-I) mRNA editing can alter genetic information at the RNA level and is known to affect protein sequence and function in eukaryotes. However, the ability of A-to-I mRNA editing to recode protein sequences in bacteria was never shown at the protein level. Furthermore, the effect of A-to-I mRNA editing itself-not by A-to-G DNA-mimicking mutations-on protein function was never demonstrated in bacteria. Here, we show at the RNA and protein levels that A-to-I mRNA editing directly recodes a tyrosine to a cysteine residue in the toxin HokB (part of the HokB/sokB toxin-antitoxin system in Escherichia coli). Consequently, the toxicity of edited HokB increases, inducing bacterial death or early entrance into the stationary phase, depending on its expression level. Furthermore, we demonstrate that in vivo disulfide bond formation underlies the effect of A-to-I mRNA editing on HokB function, suggesting that A-to-I mRNA editing constitutes a novel mechanism to regulate disulfide bond formation in bacteria. Finally, we observe that A-to-I mRNA editing of hokB is conserved in pathogenic bacteria, supporting functional importance with possible clinical relevance. Our work reveals that A-to-I mRNA editing can constitute a novel mechanism to regulate protein sequence, disulfide bonds, and function in bacteria.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
None declared.
Figures



References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

