Differential oligomerization regulates PHF13 chromatin affinity and function
- PMID: 40598901
- PMCID: PMC12214034
- DOI: 10.1093/nar/gkaf572
Differential oligomerization regulates PHF13 chromatin affinity and function
Abstract
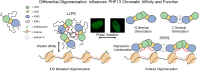
PHF13 is a H3K4me3 epigenetic reader that modulates key chromatin processes including transcription, DNA damage response, and chromatin architecture. PHF13 is found aberrantly regulated in different cancers and its misexpression alters the epigenetic landscape of key transcription factors that regulate epithelial-to-mesenchymal transition. In this study, we sought to understand how PHF13's chromatin affinity and diverse chromatin functions are intrinsically regulated. Our results show that PHF13 can oligomerize via conserved ordered regions in its N- and C- terminus increasing its chromatin valence and avidity, promoting polymer-polymer phase separation (PPPS) and chromatin inaccessibility. Impressively, a ∼3- to 5-fold overexpression of PHF13 was sufficient to globally compact chromatin visible by optical microscopy, dependent on its ordered dimerizing regions and oligomerization potential. Unexpectedly, we discovered that PHF13 can self-associate independent of its ordered domains via intrinsically disordered regions, which conversely reduced PHF13's chromatin affinity, formed liquid-liquid phase separated (LLPS) condensates, and differentially impacted gene expression. Our findings support that there is an intrinsic balance between PHF13's ordered and disordered regions and that PHF13 can phase transition between polymer-polymer and liquid-liquid phase separation states to impact chromatin structure and function.
© The Author(s) 2025. Published by Oxford University Press on behalf of Nucleic Acids Research.
Conflict of interest statement
D.H. is a founder and scientific advisor of Nuage Therapeutics.
Figures







Similar articles
-
PHF13 is a molecular reader and transcriptional co-regulator of H3K4me2/3.Elife. 2016 May 25;5:e10607. doi: 10.7554/eLife.10607. Elife. 2016. PMID: 27223324 Free PMC article.
-
Structural dynamics of IDR interactions in human SFPQ and implications for liquid-liquid phase separation.Acta Crystallogr D Struct Biol. 2025 Jul 1;81(Pt 7):357-379. doi: 10.1107/S2059798325005303. Epub 2025 Jun 27. Acta Crystallogr D Struct Biol. 2025. PMID: 40574713 Free PMC article.
-
Bath: a Bayesian approach to analyze epigenetic transitions reveals a dual role of H3K27me3 in chondrogenesis.Epigenetics Chromatin. 2025 Jun 27;18(1):38. doi: 10.1186/s13072-025-00594-6. Epigenetics Chromatin. 2025. PMID: 40571950 Free PMC article.
-
Immunogenicity and seroefficacy of pneumococcal conjugate vaccines: a systematic review and network meta-analysis.Health Technol Assess. 2024 Jul;28(34):1-109. doi: 10.3310/YWHA3079. Health Technol Assess. 2024. PMID: 39046101 Free PMC article.
-
Ear drops for the removal of ear wax.Cochrane Database Syst Rev. 2018 Jul 25;7(7):CD012171. doi: 10.1002/14651858.CD012171.pub2. Cochrane Database Syst Rev. 2018. PMID: 30043448 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

