The accessory type III secretion system effectors collectively shape intestinal inflammatory infection outcomes
- PMID: 40599135
- PMCID: PMC12233879
- DOI: 10.1080/19490976.2025.2526134
The accessory type III secretion system effectors collectively shape intestinal inflammatory infection outcomes
Abstract
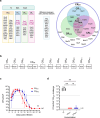
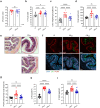
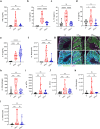
Injection of effectors via a type III secretion system (T3SS) is an infection strategy shared by various Gram-negative bacterial pathogens, many infecting mucosal surfaces. While individual T3SS effectors are well characterized, their network-level organization and the distinction between core and accessory effectors remain incompletely understood. Here, by systematically dissecting the T3SS effector network of the enteric mouse pathogen Citrobacter rodentium (CR) we identified a subset of 12 accessory effectors that, while dispensable for colonization, significantly alter infection outcomes. A strain lacking the accessory effectors (CRM12) remained virulent in susceptible mouse hosts yet resulted in reduced epithelial barrier damage, inflammation, and immune cell infiltration in resistant mice. Deep proteomic analysis specifically targeting CR-attached colonic epithelial cells revealed that, despite lacking 39% of its effector repertoire, infection with CRM12 results in similar changes to global protein expression as seen in mice infected with the wild-type strain, though key regulators of barrier integrity were differentially expressed. Using a host with impaired barrier repair (Il22- /- mice), we confirmed that accessory effectors collectively shape infection outcomes without significantly impacting virulence. This study refines the concept of core and accessory effectors, providing a basis for further studies into effector-driven host adaptation.
Keywords: Citrobacter rodentium; Type III secretion system effectors; barrier disruption; gut infection; mucosal immune responses.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




Similar articles
-
Determinants of divergent Salmonella and Shigella epithelial colonization strategies resolved in human enteroids and colonoids.mBio. 2025 Jul 9;16(7):e0091125. doi: 10.1128/mbio.00911-25. Epub 2025 May 30. mBio. 2025. PMID: 40444467 Free PMC article.
-
Edwardsiella ictaluri type III secretion system effector EseG modulates cytoskeletal dynamics and immune response in macrophages.Infect Immun. 2025 Aug 12;93(8):e0052524. doi: 10.1128/iai.00525-24. Epub 2025 Jul 10. Infect Immun. 2025. PMID: 40637600 Free PMC article.
-
Citrobacter rodentium infection activates colonic lamina propria group 2 innate lymphoid cells.PLoS Pathog. 2025 Jul 1;21(7):e1013276. doi: 10.1371/journal.ppat.1013276. eCollection 2025 Jul. PLoS Pathog. 2025. PMID: 40591723 Free PMC article.
-
Sensing for survival: specialised regulatory mechanisms of Type III secretion systems in Gram-negative pathogens.Biol Rev Camb Philos Soc. 2024 Jun;99(3):837-863. doi: 10.1111/brv.13047. Epub 2024 Jan 12. Biol Rev Camb Philos Soc. 2024. PMID: 38217090 Review.
-
How Do the Virulence Factors of Shigella Work Together to Cause Disease?Front Cell Infect Microbiol. 2017 Mar 24;7:64. doi: 10.3389/fcimb.2017.00064. eCollection 2017. Front Cell Infect Microbiol. 2017. PMID: 28393050 Free PMC article. Review.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
