Computational modeling of cotranscriptional RNA folding
- PMID: 40599241
- PMCID: PMC12212151
- DOI: 10.1016/j.csbj.2025.06.005
Computational modeling of cotranscriptional RNA folding
Abstract
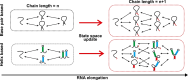
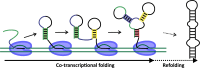
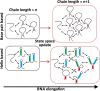
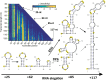
An RNA folds as it is transcribed. RNA folding during transcription differs fundamentally from thermodynamic folding. While thermodynamic folding reaches an equilibrium ensemble of structures, cotranscriptional folding is a kinetic process where the RNA structure evolves as the chain elongates during transcription. This dynamic folding pathway causes cotranscriptional structures often to deviate from thermodynamic predictions, as the system rarely reaches equilibrium. Since these cotranscriptional effects can persist in the mature RNA's structure, understanding this kinetic process is crucial. While experimental studies of cotranscriptional folding have been successful, they remain resource-intensive. Computational modeling has emerged as an increasingly practical and powerful approach for investigating these dynamics. This short review examines current computational methods and tools for simulating cotranscriptional folding, with the goal of advancing our understanding of RNA folding mechanisms.
Keywords: Computational modeling; Cotranscriptional folding; Kinetic pathway; RNA folding; Structure prediction.
© 2025 The Author(s).
Conflict of interest statement
None.
Figures
References
-
- Boyle John, Robillard Guido T. Sequential folding of transfer RNA. A nuclear magnetic resonance study of yeast phenylalanine transfer RNA. Biochemistry. 1980;19(25):6129–6136. - PubMed
-
- Pan Tao, Sosnick Tobin R. RNA folding during transcription. Annu Rev Biophys Biomol Struct. 2006;35:161–175. - PubMed
-
- Lai Daniel, Proctor Jesse R., Meyer Irmtraud M. Importance of co-transcriptional RNA folding in RNA function and stability. Structure. 2013;21(9):1439–1445.
-
- Meyer Michelle M., Breaker Ronald R. The co-transcriptional folding of riboswitches: evidence and implications. Curr Opin Struct Biol. 2004;14(3):304–310.
Publication types
LinkOut - more resources
Full Text Sources
