[One-Step Detection of Human Influenza B Virus Through Recombinase Polymerase Amplification and CRISPR/Cas12a Protein]
- PMID: 40599263
- PMCID: PMC12207055
- DOI: 10.12182/20250360105
[One-Step Detection of Human Influenza B Virus Through Recombinase Polymerase Amplification and CRISPR/Cas12a Protein]
Abstract
Objective: To establish a one-step detection method based on recombinase polymerase amplification (RPA) and CRISPR/Cas12a protein for the rapid and sensitive detection of human influenza B virus.
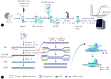
Methods: RPA amplification primers were designed according to the conserved gene (NS1 gene) of human influenza B virus (Victoria lineage). The reaction system was established using the standard plasmid as the template. First of all, the reaction system was incubated at 37 ℃ for 15 minutes for RPA amplification. Then, the CRISPR/Cas12a system on the tube cap was thoroughly mixed with the RPA amplification product at the bottom of the tube through fast centrifugation, and real-time fluorescence detection was carried out at 37 ℃. The reaction conditions were optimized to establish a one-step RPA-CRISPR/Cas12a detection method for human influenza B virus. The sensitivity of the testing method was evaluated using standard plasmids and pseudoviruses, and the specificity was evaluated using other viruses that may cause febrile respiratory syndrome. The consistency between the results of the one-step detection method and those of RT-qPCR detection was evaluated by testing real samples.
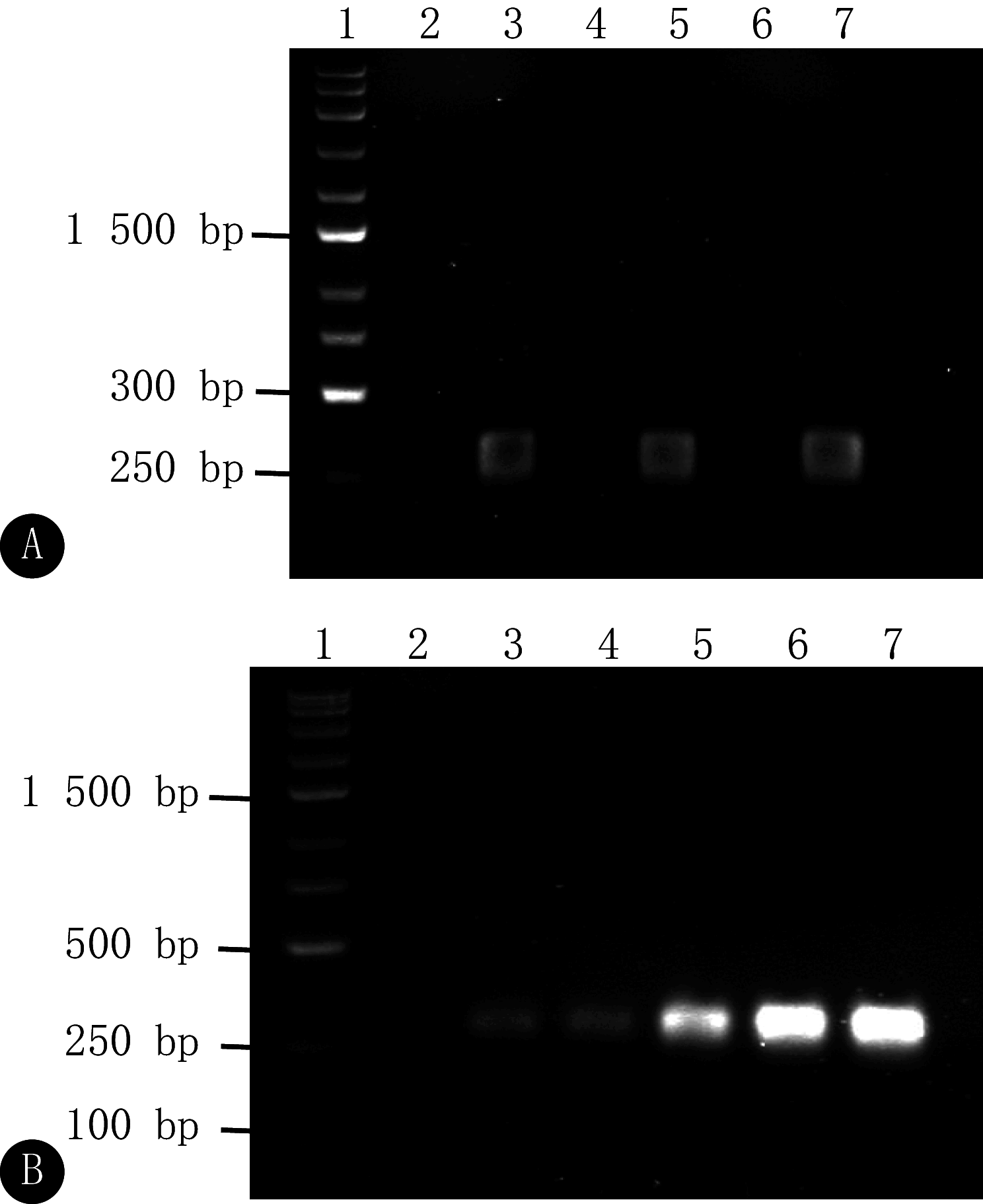
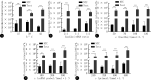
Results: A one-step detection method based on RPA-CRISPR/Cas12a was successfully established. The optimal reaction conditions included a reaction temperature of 37 ℃, a Cas12a/crRNA concentation ratio of 1∶1, a Cas12a concentration of 120 nmol/L, a single-stranded DNA (ssDNA) probe concentration of 300 nmol/L, and a primer concentration of 480 nmol/L. The method could detect standard plasmid DNA as low as 2.8 copies/μL within 25 minutes and pseudoviruses as low as 2.77 copies/μL within 30 minutes. The testing method showed high specificity, and no cross-reaction was observed with the severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), influenza A (H1N1) virus, or respiratory syncytial virus subgroup A. When testing clinical samples, the sensitivity and the specificity for examining clinical samples were 93.33% and 100%, respectively, and consistency with RT-qPCR results was 97.14%.
Conclusion: With the one-step detection method based on RPA-CRISPR/Cas12a established in this study, the whole sample detection process, including nucleic acid release, reverse transcription, isothermal amplification, CRISPR/Cas12a system cleavage, and fluorescence signal output, can be completed within 30 minutes. Its high sensitivity, specificity, and successful application in clinical samples highlight its potential for rapid point-of-care testing in clinical settings.
目的: 建立基于重组酶聚合酶扩增(recombinase polymerase amplification, RPA)和CRISPR/Cas12a蛋白的一步检测法,实现对人乙型流感病毒即时灵敏的检测。
方法: 根据人乙型流感病毒(维多利亚系)保守基因(NS1基因)设计RPA扩增引物,首先以标准质粒为模板建立反应体系,在37 ℃条件下孵育15 min进行RPA扩增;通过瞬时离心将管盖上的CRISPR/Cas12a体系与管底的RPA扩增产物充分混匀,在37 ℃进行实时荧光检测。对反应条件进行优化,建立人乙型流感病毒RPA-CRISPR/Cas12a一步检测法;以标准质粒和假病毒为模板,评价方法灵敏度;以其他发热呼吸道症候群病毒为模板,评价方法特异度;通过对真实样本的检测,评价一步检测法与RT-qPCR检测结果的一致性。
结果: 成功建立基于RPA-CRIPSR/Cas12a的一步检测法,反应温度为37 ℃,Cas12a/crRNA浓度比例为1∶1,Cas12a浓度为120 nmol/L,单链DNA(single-stranded DNA, ssDNA)探针浓度为300 nmol/L,引物浓度为480 nmol/L时最佳,25 min可检测低至2.8 copies/μL的标准质粒DNA,30 min可检测低至2.77 copies/μL的假病毒;特异性强,与新型冠状病毒、甲型流感病毒H1N1、呼吸道合胞病毒A型无交叉反应;检测真实样本的灵敏度为93.33%,特异性为100%,与RT-qPCR检测结果一致率为97.14%。
结论: 本研究建立的基于RPA-CRISPR/Cas12a的一步检测法,30 min即可完成人乙型流感病毒真实样本核酸释放、逆转录、等温扩增、CRISPR/Cas12a系统切割并输出荧光信号的完整检测流程,具有较高灵敏度和特异性,并成功应用于临床样本,在实现病毒即时检测方面具有良好的应用前景。
Keywords: CRISPR-Cas systems; Fluorimetry; Human influenza B virus; Recombinase polymerase amplification.
© 2025《四川大学学报(医学版)》编辑部 Copyright ©2025 Journal of Sichuan University (Medical Science Edition).
Conflict of interest statement
利益冲突 本文作者汪川是本刊编委会编委。该文在编辑评审过程中所有流程严格按照期刊政策进行,且未经其本人经手处理。除此之外,所有作者均声明不存在利益冲突。
Figures
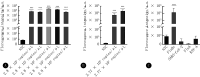
References
-
- FERRARI A J, SANTOMAURO D F, AALI A, et al Global incidence, prevalence, years lived with disability (YLDs), disability-adjusted life-years (DALYs), and healthy life expectancy (HALE) for 371 diseases and injuries in 204 countries and territories and 811 subnational locations, 1990-2021: a systematic analysis for the Global Burden of Disease Study 2021. Lancet. 2024;403(10440):2133–2161. doi: 10.1016/S0140-6736(24)00757-8. - DOI - PMC - PubMed
-
- 张勐劼, 凌益新, 黄俊驹, 等 2018-2023年莆田市乙型流感病毒流行特征和变异分析. 分子诊断与治疗杂志. 2024;16(12):2286–2290. doi: 10.19930/j.cnki.jmdt.2024.12.015. - DOI
- ZHANG M J, LING Y X, HUANG J J, et al Analysis of the prevalence characteristics and hemagglutinin gene variations of influenza type B virus in Putian City from 2018 to 2023. Journal of Molecular Diagnostics and Therapy. 2024;16(12):2286–2290. doi: 10.19930/j.cnki.jmdt.2024.12.015. - DOI
-
- 李文敏, 张娟, 刘长钰, 等 新冠疫情前后广州市甲型和乙型流感流行态势分析. 现代预防医学. 2021;48(16):3029–3032. doi: 10.20043/j.cnki.mpm.2021.16.032. - DOI
- LI W M, ZHANG J, LIU C Y, et al Epidemiological characteristics analysis of influenza A and influenza B in Guangzhou before and after COVID-19 outbreak. Modern Preventive Medicine. 2021;48(16):3029–3032. doi: 10.20043/j.cnki.mpm.2021.16.032. - DOI
-
-
US Centers for Disease Control and Prevention. About Estimated Flu Burden. (2024-11-13) [2025-03-12]. https://www.cdc.gov/flu-burden/php/about/index.html.
-
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous
