The impact of integrated genomic surveillance on non-typhoidal Salmonella infection in Australia: an ecological study
- PMID: 40599377
- PMCID: PMC12212105
- DOI: 10.1016/j.lanwpc.2025.101592
The impact of integrated genomic surveillance on non-typhoidal Salmonella infection in Australia: an ecological study
Abstract
Background: Whole Genome Sequencing (WGS) is a powerful technology for monitoring and detecting outbreaks of infectious pathogens, including non-typhoidal Salmonella (NTS). Despite its higher cost than traditional typing methods, WGS offers numerous advantages, including higher resolution and potentially quicker turnaround time. However, evidence regarding its effectiveness in NTS surveillance has predominantly stemmed from micro-simulations or small-scale data. Notably, a recent systematic review identified a lack of real-world, large-scale evidence on the impact of WGS application in NTS surveillance. Our study fills this gap by estimating the effects of WGS on NTS surveillance in Australia using national notifiable disease datasets.
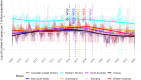
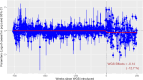
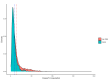
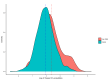
Methods: The main dataset was the National Notifiable Diseases Surveillance System (NNDSS) for NTS from 2009 to 2024. The treatment variable was defined as a binary variable representing the period when WGS was implemented in each jurisdiction of Australia. To minimise the effects of unobserved confounders, we employed a two-stage difference-in-difference (2sDiD) approach. This method estimated the parameters of state and period fixed-effects and then adjusted the observed outcomes from those fixed-effects in the first stage. The average treatment effect was obtained in the second stage by regressing the adjusted outcome against the treatment in the second stage. We also conducted a sensitivity analysis using a multi-period DiD model with a double machine-learning estimator.
Findings: Compared to the pre-WGS periods, the introduction of WGS was associated with an average of 11.6% reduction in NTS cases when a static specification was applied. Results of a dynamic specification were slightly higher, with a 12.7% reduction in NTS cases after WGS. The estimated effects increased to 17.5% when a multi-period DiD model with a double machine learning estimator was applied.
Interpretation: Our study shows that WGS was associated with a significant reduction (11.6%-17.5%) of NTS cases in Australia. Using the cost and break-even point of NTS from previous Australian studies, our findings suggest that WGS is associated with 7200-10,900 cases of NTS averted, saving US$11.3 m-US$17.0 m per year.
Funding: Australian National Health and Medical Research Council, Medical Research Futures Fund (FSPGN00049), and Investigator Grant (GNT1196103) to BPH.
Keywords: Australia; Ecological study; Genomic surveillance; Salmonella infections; Whole genome sequencing.
© 2025 The Author(s).
Conflict of interest statement
Authors declare that they have no competing interests.
Figures
References
-
- Tran M., Smurthwaite K.S., Nghiem S., et al. Economic evaluations of whole-genome sequencing for pathogen identification in public health surveillance and health-care-associated infections: a systematic review. Lancet Microbe. 2023;4(11):e953–e962. - PubMed
-
- Jain S., Mukhopadhyay K., Thomassin P.J. An economic analysis of salmonella detection in fresh produce, poultry, and eggs using whole genome sequencing technology in Canada. Food Res Int. 2019;116:802–809. - PubMed
-
- World Health Organization . World Health Organization; Geneva, Switzerland: 2015. WHO estimates of the global burden of foodborne diseases.
LinkOut - more resources
Full Text Sources
Miscellaneous

