Probiotics under Selective Pressure: Novel Insights and Biosafety Challenge
- PMID: 40599455
- PMCID: PMC12207940
- DOI: 10.32592/ARI.2024.79.6.1165
Probiotics under Selective Pressure: Novel Insights and Biosafety Challenge
Abstract
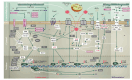
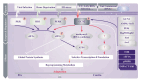
The advent of novel high-resolution physicochemical techniques and the integration of omics technologies into biomedical research have opened avenues for investigating the mechanisms underlying bacterial survival in vitro and in vivo, subjected to the influence of biotic and abiotic stressors. This encompasses axenic cultures, microbial communities, and holobionts. The development of innovative methodological platforms has facilitated the acquisition of unique data relevant to both fundamental and applied scientific fields. The experimental results indicated a remarkably high level of genomic plasticity in microorganisms and the potential for the evolution of bacterial virulence under selective pressure. These findings have significantly impacted our understanding of the arsenal of self-defense tools in bacteria and the prioritization of research in this field. The increasing quantity of factual material now necessitates a shift in focus from pathogens to the broader category of commensal bacteria, which are used as probiotics in various fields, including medicine, agriculture, and the food industry. The possibility of large-scale genomic reorganization and progressive evolution of virulence in these bacteria under stressful conditions, as well as their modulation of host cell signaling systems and suppression of innate immunity, negative regulation of key cell cycle controllers, disruption of the structure of the intestinal microbiota and intestinal homeostasis, highlight the obvious insufficiency of our knowledge about the "logic of life" of symbionts and the mechanisms of their interaction with eukaryotic cells. This may compromise the ideas of several practical applications. This underscores the importance of comprehensive studies of commensals, their potential for plasticity in different environmental conditions, and the ways in which they communicate and interact with regulatory networks of higher organisms. It also highlights the need to develop a standardization for assessing the safety of probiotics. The review addresses these issues.
Keywords: Antimicrobial Resistance; Gut Microbes; Probiotics; Selective Pressure; Virulence Evolution.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures



Similar articles
-
Synbiotics, prebiotics and probiotics for solid organ transplant recipients.Cochrane Database Syst Rev. 2022 Sep 20;9(9):CD014804. doi: 10.1002/14651858.CD014804.pub2. Cochrane Database Syst Rev. 2022. PMID: 36126902 Free PMC article.
-
Factors that influence parents' and informal caregivers' views and practices regarding routine childhood vaccination: a qualitative evidence synthesis.Cochrane Database Syst Rev. 2021 Oct 27;10(10):CD013265. doi: 10.1002/14651858.CD013265.pub2. Cochrane Database Syst Rev. 2021. PMID: 34706066 Free PMC article.
-
How lived experiences of illness trajectories, burdens of treatment, and social inequalities shape service user and caregiver participation in health and social care: a theory-informed qualitative evidence synthesis.Health Soc Care Deliv Res. 2025 Jun;13(24):1-120. doi: 10.3310/HGTQ8159. Health Soc Care Deliv Res. 2025. PMID: 40548558
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
