Python-driven impedance profiling on peptide-functionalized biosensor for detection of HIV gp41 envelope protein
- PMID: 40599495
- PMCID: PMC12209138
- DOI: 10.1007/s13205-025-04400-8
Python-driven impedance profiling on peptide-functionalized biosensor for detection of HIV gp41 envelope protein
Abstract
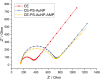
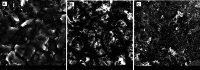
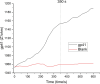
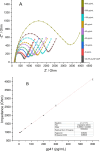
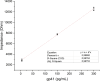
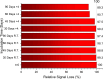
This study presents the first label-free impedimetric biosensor for the detection of HIV envelope protein gp41 using antimicrobial peptides (AMPs) as biorecognition receptors. The biosensor interface was enhanced with thiolated polystyrene and gold nanospheres to ensure stable peptide immobilization and resist nonspecific adsorption. Electrochemical impedance spectroscopy (EIS) confirmed each step of the electrode modification, while surface morphology was validated via scanning electron microscopy. A Python-based deep learning algorithm was applied to impedance data for efficient curve fitting and regression modeling. The biosensor demonstrated high sensitivity, with a linear detection range of 5-600 pg/mL, a regression coefficient (R 2) of 0.9946, a limit of detection (LOD) of 1.62 pg/mL, and a limit of quantification (LOQ) of 4.91 pg/mL. Chronoimpedimetric (CI) detection revealed that gp41 binding occurred within 350 s. The biosensor showed excellent reproducibility (CV % = 0.22%), good selectivity with less than 12% signal variation in spiked serum, and robust stability, maintaining functionality after extended storage. These results highlight the biosensor's potential as a rapid, sensitive, and reproducible diagnostic platform for early HIV detection.
Supplementary information: The online version contains supplementary material available at 10.1007/s13205-025-04400-8.
Keywords: Biosensor; Gp41; HIV; Impedance; Python.
© The Author(s) 2025.
Conflict of interest statement
Conflict of interestThe authors declare no conflict of interests.
Figures

References
-
- Conlin R, Erickson K, Abbate J, Kolemen E (2021) Keras2c: a library for converting Keras neural networks to real-time compatible C. Eng Appl Artif Intell 100:104182. 10.1016/J.ENGAPPAI.2021.104182 - DOI
-
- Dai Y, Wei J, Qin F (2024) Recurrent neural network (RNN) and long short-term memory neural network (LSTM) based data-driven methods for identifying cohesive zone law parameters of nickel-modified carbon nanotube reinforced sintered nano-silver adhesives. Mater Today Commun 39:108991. 10.1016/J.MTCOMM.2024.108991 - DOI
-
- Elalouf A, Maoz H, Rosenfeld AY (2024) Comprehensive Insights into the molecular basis of HIV glycoproteins. Appl Sci 14:8271. 10.3390/APP14188271 - DOI
LinkOut - more resources
Full Text Sources

