Structure-guided design of a methyltransferase-like 3 (METTL3) proteolysis targeting chimera (PROTAC) incorporating an indole-nicotinamide chemotype
- PMID: 40599585
- PMCID: PMC12207533
- DOI: 10.1039/d5md00359h
Structure-guided design of a methyltransferase-like 3 (METTL3) proteolysis targeting chimera (PROTAC) incorporating an indole-nicotinamide chemotype
Abstract
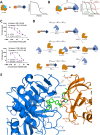
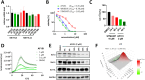
Methyltransferase-like 3 (METTL3) is the main catalytic subunit of the m6A methyltransferase complex (MTC) and plays an essential role in various disease indications, including acute myeloid leukemia (AML). Here, we describe the structure-guided design and evaluation of METTL3 proteolysis-targeting chimeras (PROTACs), starting from the potent small-molecule inhibitor STM2457. Across four design generations, we highlight key considerations, particularly regarding the exit vector, linker mechanics, and METTL3-binding chemotype composition. Our most effective PROTAC, AF151, forms a stable complex between the E3 ligase von Hippel-Lindau (VHL) and the target-of-interest METTL3, demonstrating efficient METTL3 degradation (DC50 = 430 nM) in the AML cell line MOLM-13. This molecule candidate exhibits more pronounced effects on viability inhibition (IC50 = 0.45 μM) and more significant m6A level reduction in cancer cells than its non-PRTOAC parent compounds. By incorporating the indole-nicotinamide chemotype as the METTL3-binding recruiter, this PROTAC is structurally distinct from recently published METTL3 PROTACs, expanding the design options for future METTL3 degrader development.
This journal is © The Royal Society of Chemistry.
Conflict of interest statement
There are no conflicts of interest to declare.
Figures


Similar articles
-
A close-up shot of protein-protein docking, from experiment to theory and reverse with the PROTAC performers.J Biomol Struct Dyn. 2025 Jul;43(11):5260-5267. doi: 10.1080/07391102.2024.2308778. Epub 2024 Jan 29. J Biomol Struct Dyn. 2025. PMID: 38284361
-
Unveiling New Triazoloquinoxaline-Based PROTACs Designed for the Selective Degradation of the ncBAF Chromatin Remodeling Subunit BRD9.Chemistry. 2025 Jun 17;31(34):e202404218. doi: 10.1002/chem.202404218. Epub 2025 May 20. Chemistry. 2025. PMID: 40343767 Free PMC article.
-
Hijacking the MDM2 E3 Ligase with Novel BRD4-Targeting Proteolysis-Targeting Chimeras in Pancreatic Cancer Cells.Chembiochem. 2025 Jul 11;26(13):e202500133. doi: 10.1002/cbic.202500133. Epub 2025 Jun 23. Chembiochem. 2025. PMID: 40317844 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources

