Novel potential biomarkers for predicting childhood caries via metagenomic analysis
- PMID: 40599650
- PMCID: PMC12209247
- DOI: 10.3389/fcimb.2025.1522970
Novel potential biomarkers for predicting childhood caries via metagenomic analysis
Abstract
Background: Dental caries is a prevalent global health issue, particularly among children, with significant oral and overall health implications. The oral microbiome is considered a critical factor in caries development, with various microbial species implicated in the disease process.
Objectives: This study aims to explore the changes and interactions of oral microbiota in childhood caries using metagenomic analysis, and identify potential biomarkers for early caries detection and treatment.
Methods: Saliva samples were collected from 241 children aged 6 to 9 years, categorized into caries-free (CF), low-caries (CL), and caries-severe (CS) groups. Metagenomic sequencing was performed to analyze the oral microbiome, followed by a series of statistical and functional analyses to characterize microbial diversity and function.
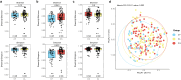
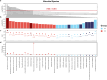
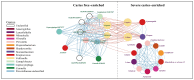
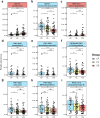
Results: The study revealed significant differences in the microbial community composition among the groups, with the CS group exhibiting higher alpha and beta diversity than that of the CF group. Numerous unclassified microorganisms, such as Campylobacter SGB19347 and Catonella SGB4501, are intimately linked to dental caries and display intricate interaction networks, suggesting the potential formation of a distinct ecological network. In functional assessment, we identified a possible link between pectin and caries, suggesting that microorganisms that produce pectinase enzymes might play a role in the advancement of severe dental caries. Additionally, we identified 16 species as the best marker for severe dental caries, achieving an impressive AUC of 0.91.
Conclusion: The role of microbiota in dental caries is multifaceted, involving a complex interplay of microbial species and functions. Our findings enhance the understanding of the microbial basis of dental caries and offer potential diagnostic and therapeutic targets. The predictive capacity of the identified biomarkers warrants further investigation for early caries detection and intervention.
Clinical significance: The identification of novel biomarkers through metagenomic analysis enables early detection and targeted intervention for childhood caries, potentially transforming children dental care and significantly improving long-term oral health outcomes.
Keywords: biomarkers; children; dental caries; metagenomic sequencing; oral microbiome.
Copyright © 2025 Zhang, Zheng, Huang, Zou, Zhang, Repo, Yin, You, Jie and Xu.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

Similar articles
-
Caries influences the composition of oral microorganisms in male children with ASD.Microb Pathog. 2025 Oct;207:107904. doi: 10.1016/j.micpath.2025.107904. Epub 2025 Jul 14. Microb Pathog. 2025. PMID: 40669761
-
Atraumatic restorative treatment versus conventional restorative treatment for managing dental caries.Cochrane Database Syst Rev. 2017 Dec 28;12(12):CD008072. doi: 10.1002/14651858.CD008072.pub2. Cochrane Database Syst Rev. 2017. PMID: 29284075 Free PMC article.
-
Ensemble learning for microbiome-based caries diagnosis: multi-group modeling and biological interpretation from salivary and plaque metagenomic data.BMC Oral Health. 2025 Jul 17;25(1):1188. doi: 10.1186/s12903-025-06590-2. BMC Oral Health. 2025. PMID: 40676575 Free PMC article.
-
WITHDRAWN: Dental fillings for the treatment of caries in the primary dentition.Cochrane Database Syst Rev. 2016 Oct 17;10(10):CD004483. doi: 10.1002/14651858.CD004483.pub3. Cochrane Database Syst Rev. 2016. PMID: 27748505 Free PMC article.
-
Dental fillings for the treatment of caries in the primary dentition.Cochrane Database Syst Rev. 2009 Apr 15;(2):CD004483. doi: 10.1002/14651858.CD004483.pub2. Cochrane Database Syst Rev. 2009. Update in: Cochrane Database Syst Rev. 2016 Oct 17;10:CD004483. doi: 10.1002/14651858.CD004483.pub3. PMID: 19370602 Updated.
References
-
- Al-Hebshi N. N., Baraniya D., Chen T., Hill J., Puri S., Tellez M., et al. (2019). Metagenome sequencing-based strain-level and functional characterization of supragingival microbiome associated with dental caries in children. J. Microbiol 11, 1557986. doi: 10.1080/20002297.2018.1557986 - DOI - PMC - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

