Nuclear factor IA-mediated transcriptional regulation of crystallin αB inhibits hepatocellular carcinoma progression
- PMID: 40599718
- PMCID: PMC12210110
- DOI: 10.3892/mco.2025.2867
Nuclear factor IA-mediated transcriptional regulation of crystallin αB inhibits hepatocellular carcinoma progression
Abstract
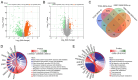
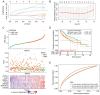
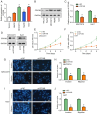
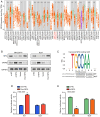
Hepatocellular carcinoma (HCC) is a highly invasive malignant tumor with limited therapeutic options. In the present study, bioinformatics analysis, including differential expression analysis, functional enrichment, protein-protein interaction network construction, survival analysis and risk model evaluation, identified CRYAB as a central prognostic gene in HCC. Additionally, motif analysis using JASPAR revealed nuclear factor IA (NFIA), as a potential transcriptional regulator of CRYAB. Further in vitro experiments were conducted to explore the roles of CRYAB and NFIA in HCC, suggesting that these molecules may serve as promising therapeutic targets for future research. Differentially expressed genes (DEGs) from the Cancer Genome Atlas-liver hepatocellular carcinoma (LIHC) and GSE113996 datasets were identified using the 'limma' package, with Biological Process and Kyoto Encyclopedia of Genes and Genomes enrichment analysis conducted. Overlapping DEGs underwent Protein-protein interaction and prognostic analysis. Key prognostic genes were selected through Kaplan-Meier survival analysis and Least Absolute Shrinkage and Selection Operator regression before they were incorporated into a predictive risk model, which was evaluated by receiver operating characteristic analysis. JASPAR motif analysis identified NFIA as a potential transcriptional regulator of CRYAB, with the TIMER database used to further examine the NFIA expression profile among other cancers. In vitro assays using MHCC97H and Huh7 cells were used to examine the roles of CRYAB and NFIA in HCC. Cell counting kit-8 (CCK-8) assay was used to assess proliferation, whilst Transwell assay was used to measure migration and invasion. To investigate the reciprocal regulation, rescue experiments combining NFIA overexpression and CRYAB knockdown were performed to compare their effects on cell proliferation, migration and invasion. Additionally, dual-luciferase assay was used to examine the regulatory effect of NFIA on the CRYAB promoter by comparing the wild-type and mutant constructs. Bioinformatics analyses revealed CRYAB to be a hub gene. CRYAB upregulation was found to be associated with poor prognosis in patients with LIHC. In vitro, elevated CRYAB expression was observed in HCC cell lines compared with that in the huma liver immortalized cell line THLE-2. CRYAB knockdown was found to significantly inhibit MHCC97H and Huh7 cell proliferation, migration and invasion. By contrast, NFIA expression was found to be downregulated in LIHC compared with that in normal liver tissues, where its expression showed an inverse association with that of CRYAB. Direct interaction between NFIA and the CRYAB promoter region was confirmed through dual-luciferase assays. Furthermore, low NFIA expression markedly enhanced HCC cell proliferation, invasion and migration. This pro-tumor effect was reversed in the si-NFIA + si-CRYAB group, where simultaneous downregulation of CRYAB significantly reduced cell proliferation, migration and invasion, suggesting that CRYAB downregulation can counteract the effects induced by low NFIA expression. To conclude, these results suggest that NFIA can inhibit the malignant proliferation of HCC cells by activating CRYAB expression, which further suggest that CRYAB and NFIA are promising avenues for the development of novel HCC treatment strategies.
Keywords: crystallin αB; hepatocellular carcinoma; in vitro evaluation; nuclear factor IA; transcriptional regulation.
Copyright: © 2025 Jin et al.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
A novel glutamine metabolism-related risk model for prognostic prediction of liver hepatocellular carcinoma.Oncol Lett. 2025 Jun 20;30(3):403. doi: 10.3892/ol.2025.15149. eCollection 2025 Sep. Oncol Lett. 2025. PMID: 40613031 Free PMC article.
-
Caveolin-1 inhibits the proliferation and invasion of lung adenocarcinoma via EGFR degradation.Sci Rep. 2025 Jul 1;15(1):21654. doi: 10.1038/s41598-025-05259-8. Sci Rep. 2025. PMID: 40594106 Free PMC article.
-
Bioinformatics identification and validation of m6A/m1A/m5C/m7G/ac4 C-modified genes in oral squamous cell carcinoma.BMC Cancer. 2025 Jul 1;25(1):1055. doi: 10.1186/s12885-025-14216-7. BMC Cancer. 2025. PMID: 40597017 Free PMC article.
-
Cost-effectiveness of using prognostic information to select women with breast cancer for adjuvant systemic therapy.Health Technol Assess. 2006 Sep;10(34):iii-iv, ix-xi, 1-204. doi: 10.3310/hta10340. Health Technol Assess. 2006. PMID: 16959170
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
References
-
- Hahn JW, Woo S, Park J, Lee H, Kim HJ, Ko JS, Moon JS, Rahmati M, Smith L, Jang J, et al. Global, Regional, and National Trends in Liver Disease-Related Mortality Across 112 Countries From 1990 to. 2021, With Projections to 2050: Comprehensive Analysis of the WHO Mortality Database. J Korean Med Sci. 2024;39(e292) doi: 10.3346/jkms.2024.39.e292. - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
