Identification of potential prognostic biomarkers of thymoma with myasthenia gravis based on serum proteomics
- PMID: 40599774
- PMCID: PMC12212110
- DOI: 10.3389/fimmu.2025.1580219
Identification of potential prognostic biomarkers of thymoma with myasthenia gravis based on serum proteomics
Erratum in
-
Correction: Identification of potential prognostic biomarkers of thymoma with myasthenia gravis based on serum proteomics.Front Immunol. 2025 Jul 9;16:1650718. doi: 10.3389/fimmu.2025.1650718. eCollection 2025. Front Immunol. 2025. PMID: 40703512 Free PMC article.
Abstract
Background: Thymoma is often associated with myasthenia gravis (MG), and the resection of thymoma improves myasthenic symptoms in patients with thymoma and MG (TMG), but some patients still have no relief. Through proteomic analysis, we examined preoperative serum samples from patients with TMG to identify key prognostic proteins that could serve as a foundation for clinically predicting postoperative efficacy and guiding treatment selection.
Method: According to the Clinical Research Guidelines of the Myasthenia Gravis Foundation of America (MGFA) for Post-Intervention Status (PIS), 20 patients with TMG were divided into an effective group (T1) [the PIS was minimal manifestation status (MMS) and above] and an ineffective group (T2) (the PIS did not reach MMS and above), with 10 cases each, and a healthy control group (C) with nine cases. Blood samples from the three groups were collected through data-independent acquisition (DIA) proteomic analysis performed by mass spectrometry to identify differentially expressed proteins and search for key proteins associated with myasthenia prognosis. Finally, the target proteins were validated through the utilization of enzyme-linked immunosorbent assay (ELISA).
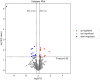
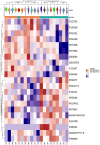
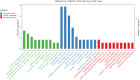
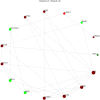
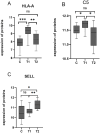
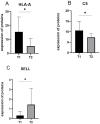
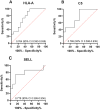
Results: A total of 514 proteins were identified in this research. Between the T1 and T2 groups, there were 20 proteins that exhibited differential expression, with 10 showing upregulation and 10 displaying downregulation. The Kyoto Encyclopedia of Genes and Genomes (KEGG) functional annotation indicated that these proteins were mainly involved in signaling pathways such as complement and coagulation cascade, prion disease, systemic lupus erythematosus, neutrophil extracellular trap formation, and transcription dysregulation in cancer. Three proteins were discovered to have a significant correlation with the prognosis of TMG: L-selectin (SELL) was downregulated, and human leukocyte antigen (HLA) class I histocompatibility antigen (HLA-A) and complement 5 (C5) were upregulated. ELISA results confirmed the proteomic results.
Conclusion: HLA-A, C5, and SELL may be potential prognostic biomarkers of TMG. This study may provide a more accurate prognostic risk assessment of TMG patients to help clinicians better individualize the initial treatment regimen for patients with different risk stratification, plan a more reasonable frequency of follow-up visits, and make more precise maintenance treatment decisions, thereby improving the overall prognosis of TMG patients.
Keywords: biomarkers; myasthenia gravis; prognostic; proteomics; thymoma.
Copyright © 2025 Lin, Liu and Qi.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

