Identification of anoikis-related subtypes and a risk score prognosis model, the association with TME landscapes and therapeutic responses in hepatocellular carcinoma
- PMID: 40599775
- PMCID: PMC12209314
- DOI: 10.3389/fimmu.2025.1602831
Identification of anoikis-related subtypes and a risk score prognosis model, the association with TME landscapes and therapeutic responses in hepatocellular carcinoma
Abstract
Introduction: Anoikis is a distinct form of programmed cell death, differing from classical apoptosis, and its role in malignant tumor progression, particularly in hepatocellular carcinoma (HCC), remains insufficiently understood. This study aims to elucidate the prognostic significance and therapeutic relevance of anoikis-related genes (ARGs) in HCC.
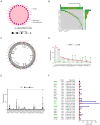
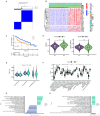
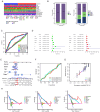
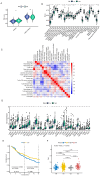
Methods: We systematically analyzed the expression, mutation, and copy number variation profiles of 27 known ARGs in HCC using public datasets. Unsupervised consensus clustering was performed to classify patients into anoikis subtypes. Weighted Gene Co-expression Network Analysis (WGCNA) identified hub gene modules, and LASSO Cox regression was applied to construct a prognostic risk score model. Correlations between the risk model and clinical outcomes, tumor microenvironment (TME) characteristics, and immunotherapy responses were evaluated. Single-cell RNA-seq and pan-cancer analyses were conducted to explore gene expression across cell types and cancer types. Finally, in vitro experiments were performed to validate the biological function of model genes.
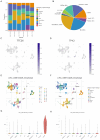
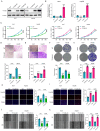
Results: Two distinct anoikis subtypes with differing prognoses and TME features were identified in HCC. A two-gene prognostic model (TTC26 and TPX2) was developed, demonstrating robust performance in predicting patient outcomes. High-risk patients exhibited lower overall survival and distinct immune infiltration profiles. Pan-cancer analysis showed widespread dysregulation of TTC26 and TPX2. In vitro experiments confirmed that TTC26 promotes HCC cell proliferation, migration, and invasion.
Discussion: Our findings reveal that anoikis-related molecular classification is closely linked to HCC prognosis and immune landscape. The established prognostic model has potential clinical utility for risk stratification and treatment guidance. TTC26 may serve as a novel biomarker and therapeutic target in HCC.
Keywords: anoikis; hepatocellular carcinoma; immunotherapy response; prognostic signature; tumor microenvironment.
Copyright © 2025 Zhai, Li, Ding, Wu, Zhang, Zhang, Zhou, Liu, Zhang and Jin.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures


Similar articles
-
Characterization of novel anoikis-related genes as prognostic biomarkers and key determinants of the immune microenvironment in esophageal cancer.Front Immunol. 2025 Jul 11;16:1599171. doi: 10.3389/fimmu.2025.1599171. eCollection 2025. Front Immunol. 2025. PMID: 40746552 Free PMC article.
-
A Novel RNA Methylation-Related Prognostic Signature and its Tumor Microenvironment Characterization in Hepatocellular Carcinoma.Technol Cancer Res Treat. 2024 Jan-Dec;23:15330338241276895. doi: 10.1177/15330338241276895. Technol Cancer Res Treat. 2024. PMID: 39155614 Free PMC article.
-
Construction and validation of a lipid metabolism-related genes prognostic signature for skin cutaneous melanoma.Biochem Biophys Res Commun. 2025 Aug 15;775:152115. doi: 10.1016/j.bbrc.2025.152115. Epub 2025 May 29. Biochem Biophys Res Commun. 2025. PMID: 40460484
-
Deciphering adenosine signaling in hepatocellular carcinoma: Pathways, prognostic models, and therapeutic implications.Clin Mol Hepatol. 2025 Jul;31(3):706-729. doi: 10.3350/cmh.2024.1068. Epub 2025 Feb 5. Clin Mol Hepatol. 2025. PMID: 39905839 Free PMC article. Review.
-
Ablative and non-surgical therapies for early and very early hepatocellular carcinoma: a systematic review and network meta-analysis.Health Technol Assess. 2023 Dec;27(29):1-172. doi: 10.3310/GK5221. Health Technol Assess. 2023. PMID: 38149643 Free PMC article.
References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

