Subclinical ketosis in postpartum dairy cows alters the adipose tissue immunological profile in a depot-specific manner
- PMID: 40599780
- PMCID: PMC12212114
- DOI: 10.3389/fimmu.2025.1578669
Subclinical ketosis in postpartum dairy cows alters the adipose tissue immunological profile in a depot-specific manner
Abstract
Introduction: Subclinical ketosis (SCK) is a common metabolic disorder linked to adipose tissue (AT) dysfunction in periparturient dairy cows. While subcutaneous AT (SAT) and visceral AT (VAT) differ in structure, cellularity, and function, depot-specific responses to ketosis remain poorly understood. This study aimed to determine the transcriptional differences of flank SAT and omental VAT in early lactation dairy cows in response to SCK.
Methods: Multiparous Holstein dairy cows within the first 10 days postpartum were screened for SCK. Subclinical ketosis was defined as blood β-hydroxybutyrate (BHB) concentrations between 1.4 and 2.6 mmol/L, while control, non-ketotic animals (NK) had BHB equal to or lower than 0.8 mmol/L. Adipose tissue biopsies were obtained from flank SAT and omental VAT from five SCK and five NK cows for RNA sequencing and immunohistochemistry analyses.
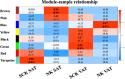
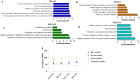
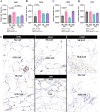
Results and discussion: Subclinical ketosis affected AT transcriptional profiles in a depot-specific manner. In SAT, transcriptional changes related to SCK reflected homeostatic AT remodeling and immune cell infiltration indicative of inflammatory responses, fibroplasia, and the negative regulation of adaptive immunity responses. In VAT, SCK-related transcriptional changes reflected increased pro-inflammatory responses linked to impaired lipid metabolism and dysregulation of focal adhesion and endocytosis. Tissue expression of proteins coded by genes differentially expressed between SCK and NK revealed a depot-dependent response on AT, indicating a higher infiltration of macrophages and B cells in SCK cows. Overall, our study provides new insights into molecular mechanisms underlying ketosis pathogenesis, highlighting the dysregulation of inflammatory responses, lipid metabolism, and insulin signaling in both SAT and VAT of postpartum dairy cows.
Keywords: immune response; ketosis; subcutaneous adipose tissue; transcriptome; visceral adipose tissue.
Copyright © 2025 Michelotti, Menarim, Tegeler, Fiallo Diez, Machado, Jones-Hall, Benitez, Loux and Strieder-Barboza.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures





References
-
- Overton TR, Waldron MR. Nutritional management of transition dairy cows: stratagies to optimize metabolic health. J Dairy Cows. (2004) 87:E105–E19. doi: 10.3168/jds.S0022-0302(04)70066-1 - DOI
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Research Materials

