Investigating Overlapping Genetic Factors and Novel Causal Genes in Autoimmune Diseases: A Transcriptome-Wide Association and Multiomics Study
- PMID: 40599891
- PMCID: PMC12213038
- DOI: 10.1155/ijog/9595651
Investigating Overlapping Genetic Factors and Novel Causal Genes in Autoimmune Diseases: A Transcriptome-Wide Association and Multiomics Study
Abstract
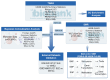
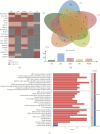
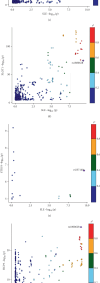
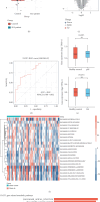
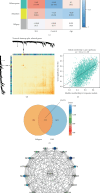
Background: Autoimmune diseases exhibit familial clustering and co-occurrence, suggesting the presence of shared genetic risk factors. However, the overlapping genetic factors across these diseases have yet to be fully elucidated. This study aimed to identify shared genetic factors across five autoimmune diseases: systemic lupus erythematosus (SLE), rheumatoid arthritis (RA), ankylosing spondylitis (AS), Sjögren's syndrome (SS), and polymyalgia rheumatica (PMR). Methods: A blood tissue-based transcriptome-wide association study (TWAS) was conducted to identify candidate genes. Bayesian colocalization analysis was employed to pinpoint genetic variants shared across diseases. Multiomics summary data-based Mendelian randomization (SMR) was used to identify causal risk genes, while transcriptomic analysis, gene set variation analysis (GSVA), and weighted gene coexpression network analysis (WGCNA) were applied to further investigate the functional roles of these genes. Results: The TWAS identified 78 candidate genes across the five autoimmune diseases. Bayesian colocalization analysis revealed five genes, GTF2H4, FLOT1, HCP5, IER3, and STK19, that share genetic variants across these disorders. Specifically, RA and AS shared independent variants of GTF2H4 (rs2230365 and rs147708689, respectively). HCP5 variants were shared with SS (rs1800628) and SLE (rs1150757), and rs1800628 was also identified as a shared locus in FLOT1 for SLE. SMR analysis highlighted FLOT1 as a strong causal risk gene for SLE. Transcriptomic analysis showed that FLOT1 is highly expressed in T cells and platelets, with involvement in multiple metabolic pathways. WGCNA identified four key neighboring genes, EHD1, SLC10A3, LMNA, and STXBP2, associated with FLOT1. Conclusion: This study uncovers shared genetic factors across five autoimmune diseases, with FLOT1 identified as a novel causal risk gene for SLE. These findings suggest that platelet-mediated pathogenic mechanisms may contribute to SLE, providing a potential target for future therapeutic interventions.
Keywords: SLE; autoimmune disease; flotillin-1 (FLOT1); platelet; summary data–based Mendelian randomization (SMR).
Copyright © 2025 Leihua Fu et al. International Journal of Genomics published by John Wiley & Sons Ltd.
Conflict of interest statement
The authors declare no conflicts of interest.
Figures

Similar articles
-
Genetic Causal Relationship Between Systemic Lupus Erythematosus and Malignant Tumors of the Female Reproductive System: A GWAS Analysis in European Populations.Hum Mutat. 2025 May 15;2025:7447886. doi: 10.1155/humu/7447886. eCollection 2025. Hum Mutat. 2025. PMID: 40406544 Free PMC article.
-
Genetic association between STAT1 and systemic lupus erythematosus: A two-sample Mendelian randomization observational study.Medicine (Baltimore). 2025 May 30;104(22):e42593. doi: 10.1097/MD.0000000000042593. Medicine (Baltimore). 2025. PMID: 40441224 Free PMC article.
-
Palmitoylation Dynamics in Systemic Lupus Erythematosus: Multi-Omics Insights and Potential Therapeutic Implications.Int J Rheum Dis. 2025 Jul;28(7):e70346. doi: 10.1111/1756-185X.70346. Int J Rheum Dis. 2025. PMID: 40613570
-
Signs and symptoms to determine if a patient presenting in primary care or hospital outpatient settings has COVID-19.Cochrane Database Syst Rev. 2022 May 20;5(5):CD013665. doi: 10.1002/14651858.CD013665.pub3. Cochrane Database Syst Rev. 2022. PMID: 35593186 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

