miR-107 represses DMPK and is sequestered by CUG repeats triggering the MSI2/miR-7 pathogenesis axis in myotonic dystrophy
- PMID: 40599975
- PMCID: PMC12212150
- DOI: 10.1016/j.omtn.2025.102584
miR-107 represses DMPK and is sequestered by CUG repeats triggering the MSI2/miR-7 pathogenesis axis in myotonic dystrophy
Abstract
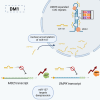
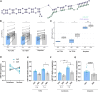
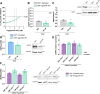
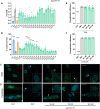
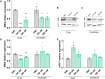
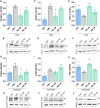
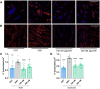
Myotonic dystrophy type 1 (DM1) is a multisystem genetic disorder characterized by muscle disease, including muscle atrophy partially originating from excessive autophagy. We have previously demonstrated that excessive Musashi-2 (MSI2) repressed the biogenesis of miR-7, which derepressed autophagy, ultimately contributing to muscle atrophy, but the root cause of MSI2 dysregulation is unknown. Herein, we investigate the intricate role of miR-107 in DM1 pathogenesis, focusing on its involvement in the MSI2>miR-7>autophagy axis as this microRNA (miRNA) directly regulates MSI2. We found that in DM1, miR-107 function is impaired because expanded CUG repeats sequester it, causing an increase in the expression of its targets, including MSI2. Through different experimental approaches, including luciferase reporter assays, differential scanning fluorimetry, and electrophoretic mobility shift assay (EMSA), we confirm that miR-107 directly binds to CUG repeats in mutant DMPK transcripts. DMPK posttranscriptional regulation by miR-107 was also demonstrated. Modulation of miR-107 in a DM1 cell model context significantly affects its downstream targets, MSI2 and miR-7, thus decreasing excessive autophagic markers and restoring pathological phenotypes such as ribonuclear foci and impaired fusion capacity. These findings underscore the critical role of miR-107 in regulating the MSI2>miR-7>autophagy axis and support this miRNA as a promising therapeutic target for correcting muscle dysfunction in DM1.
Keywords: ASO; CUG repeats; DM1; DMPK; MSI2; MT: Oligonucleotides: Therapies and Applications; miR-107; miRNA; muscle atrophy.
© 2025 The Author(s).
Conflict of interest statement
The authors declare no competing interests.
Figures


References
-
- Brook J.D., McCurrach M.E., Harley H.G., Buckler A.J., Church D., Aburatani H., Hunter K., Stanton V.P., Thirion J.P., Hudson T., et al. Molecular basis of myotonic dystrophy: expansion of a trinucleotide (CTG) repeat at the 3’ end of a transcript encoding a protein kinase family member. Cell. 1992;68:799–808. doi: 10.1016/0092-8674(92)90154-5. - DOI - PubMed
LinkOut - more resources
Full Text Sources

