Biases and complementarity in gut viromes obtained from bulk and virus-like particle-enriched metagenomic sequencing
- PMID: 40600714
- PMCID: PMC12323601
- DOI: 10.1128/spectrum.00013-25
Biases and complementarity in gut viromes obtained from bulk and virus-like particle-enriched metagenomic sequencing
Abstract
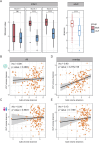
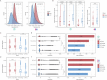
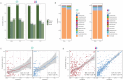
Due to varying sequencing strategies, current gut virome findings show significant variability. Specifically, bulk- and virus-like particle (VLP)-enriched metagenomic sequencing (termed bulk and VLP, respectively) present unique advantages and limitations, affecting viral genome discovery, taxonomic annotation, and community structure analysis. A comprehensive comparison of these strategies is crucial for thoroughly understanding the gut virome. This study comprehensively compared gut viromes identified from paired bulk and VLP data from 151 adult and 141 infant fecal samples. The VLP method showed superior performance to bulk in viral genome discovery in both data sets by recovering longer and more complete viral genomes, with higher sensitivity for low-abundant ones, resulting in a higher taxonomic annotation rate. However, we observed no correlations in the viral community structure (i.e., Shannon diversities) between bulk- and VLP-derived viromes, implying biases introduced during VLP enrichment. Such biases could be caused by the bacterial host features, such as the structural differences in cell walls and the prevalence and abundance of the viruses. Viruses that are of low prevalence, low abundance, or have Gram-positive bacteria as their hosts were enriched in VLP-derived viromes, in both the adult and infant data sets. Significant complementarity was observed between bulk and VLP viromes, with only about a quarter (26.7% in infants; 29.3% in adults) of VLP-viral genomes overlapping with bulk viruses. Together, our study identifies causal factors underlying the biases of bulk and VLP strategies in human gut virome studies and advocates the use of both strategies to enhance a comprehensive understanding of gut viromes.
Importance: The two mainstream gut phageome profiling strategies, namely bulk and virus-like particle (VLP), generated significantly overlapped results and have their own merits and drawbacks. Particularly, VLP exhibits higher efficiency in obtaining more, longer, and more complete viral genomes. However, VLP sequencing has the potential to alter the natural structure of viral communities, often resulting in the identification of viruses with lower prevalence and those specifically associated with Gram-positive bacterial hosts. While bulk metagenome features a more stable and diverse community, which can well reveal the interactions between viruses and bacteria. Nevertheless, bulk sequencing can suffer from lower coverage, leading to fragmented sequences and potentially missing some viral species. Therefore, it is essential to recognize that these methods are complementary rather than competitive in the comprehensive characterization of the gut phageome.
Keywords: bulk-metagenomic sequencing; gut health; gut phage; gut virus; viral-like particle metagenomic sequencing; virome.
Conflict of interest statement
The authors declare no conflict of interest.
Figures


Similar articles
-
Viromes vs. mixed community metagenomes: choice of method dictates interpretation of viral community ecology.Microbiome. 2024 Oct 7;12(1):195. doi: 10.1186/s40168-024-01905-x. Microbiome. 2024. PMID: 39375774 Free PMC article.
-
Enrichment Culture but Not Metagenomic Sequencing Identified a Highly Prevalent Phage Infecting Lactiplantibacillus plantarum in Human Feces.Microbiol Spectr. 2023 Jun 15;11(3):e0434022. doi: 10.1128/spectrum.04340-22. Epub 2023 Mar 30. Microbiol Spectr. 2023. PMID: 36995238 Free PMC article.
-
The salivary virome during childhood dental caries.mSphere. 2025 Jul 28:e0019825. doi: 10.1128/msphere.00198-25. Online ahead of print. mSphere. 2025. PMID: 40719460
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
[Volume and health outcomes: evidence from systematic reviews and from evaluation of Italian hospital data].Epidemiol Prev. 2013 Mar-Jun;37(2-3 Suppl 2):1-100. Epidemiol Prev. 2013. PMID: 23851286 Italian.
References
-
- Stockdale SR, Shkoporov AN, Khokhlova EV, Daly KM, McDonnell SA, O’ Regan O, Nolan JA, Sutton TDS, Clooney AG, Ryan FJ, Sheehan D, Lavelle A, Draper LA, Shanahan F, Ross RP, Hill C. 2023. Interpersonal variability of the human gut virome confounds disease signal detection in IBD. Commun Biol 6:221. doi: 10.1038/s42003-023-04592-w - DOI - PMC - PubMed
-
- Nishijima S, Nagata N, Kiguchi Y, Kojima Y, Miyoshi-Akiyama T, Kimura M, Ohsugi M, Ueki K, Oka S, Mizokami M, Itoi T, Kawai T, Uemura N, Hattori M. 2022. Extensive gut virome variation and its associations with host and environmental factors in a population-level cohort. Nat Commun 13:5252. doi: 10.1038/s41467-022-32832-w - DOI - PMC - PubMed
MeSH terms
Grants and funding
- Nos. 32070660/National Natural Science Foundation of China
- T2225015,61932008/National Natural Science Foundation of China
- 82161138017/NNSF-VR Sino-Swedish Joint Research Programme
- 2019YFA0905600/National Key Research and Development Program of China
- 2020YFA0712403/National Key Research and Development Program of China
LinkOut - more resources
Full Text Sources

