Leveraging protein language models for cross-variant CRISPR/Cas9 sgRNA activity prediction
- PMID: 40600900
- PMCID: PMC12254127
- DOI: 10.1093/bioinformatics/btaf385
Leveraging protein language models for cross-variant CRISPR/Cas9 sgRNA activity prediction
Abstract
Motivation: Accurate prediction of single-guide RNA (sgRNA) activity is crucial for optimizing the CRISPR/Cas9 gene-editing system, as it directly influences the efficiency and accuracy of genome modifications. However, existing prediction methods mainly rely on large-scale experimental data of a single Cas9 variant to construct Cas9 protein (variants)-specific sgRNA activity prediction models, which limits their generalization ability and prediction performance across different Cas9 protein (variants), as well as their scalability to the continuously discovered new variants.
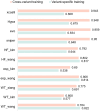
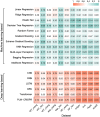
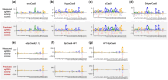
Results: In this study, we proposed PLM-CRISPR, a novel deep learning-based model that leverages protein language models to capture Cas9 protein (variants) representations for cross-variant sgRNA activity prediction. PLM-CRISPR uses tailored feature extraction modules for both sgRNA and protein sequences, incorporating a cross-variant training strategy and a dynamic feature fusion mechanism to effectively model their interactions. Extensive experiments demonstrate that PLM-CRISPR outperforms existing methods across datasets spanning seven Cas9 protein (variants) in three real-world scenarios, demonstrating its superior performance in handling data-scarce situations, including cases with few or no samples for novel variants. Comparative analyses with traditional machine learning and deep learning models further confirm the effectiveness of PLM-CRISPR. Additionally, motif analysis reveals that PLM-CRISPR accurately identifies high-activity sgRNA sequence patterns across diverse Cas9 protein (variants). Overall, PLM-CRISPR provides a robust, scalable, and generalizable solution for sgRNA activity prediction across diverse Cas9 protein (variants).
Availability and implementation: The source code can be obtained from https://github.com/CSUBioGroup/PLM-CRISPR.
© The Author(s) 2025. Published by Oxford University Press.
Figures


Similar articles
-
Harnessing an anti-CRISPR protein for powering CRISPR/Cas9-mediated genome editing in undomesticated Bacillus strains.Microb Cell Fact. 2025 Jun 23;24(1):143. doi: 10.1186/s12934-025-02776-z. Microb Cell Fact. 2025. PMID: 40551141 Free PMC article.
-
Modulating binding affinity of aptamer-based loading constructs enhances extracellular vesicle-mediated CRISPR/Cas9 delivery.J Control Release. 2025 Aug 10;384:113853. doi: 10.1016/j.jconrel.2025.113853. Epub 2025 May 18. J Control Release. 2025. PMID: 40393529
-
Focused ultrasound and microbubble-mediated delivery of CRISPR-Cas9 ribonucleoprotein to human induced pluripotent stem cells.Mol Ther. 2025 Mar 5;33(3):986-996. doi: 10.1016/j.ymthe.2025.01.013. Epub 2025 Jan 10. Mol Ther. 2025. PMID: 39797397
-
Artificial Intelligence in CRISPR-Cas Systems: A Review of Tool Applications.Methods Mol Biol. 2025;2952:243-257. doi: 10.1007/978-1-0716-4690-8_14. Methods Mol Biol. 2025. PMID: 40553337 Review.
-
CRISPR/Cas9-mediated genome editing in Ganoderma lucidum: recent advances and biotechnological opportunities.World J Microbiol Biotechnol. 2025 Jun 25;41(7):223. doi: 10.1007/s11274-025-04458-9. World J Microbiol Biotechnol. 2025. PMID: 40560492 Review.
Cited by
-
2OMe-LM: predicting 2'-O-methylation sites in human RNA using a pre-trained RNA language model.Bioinformatics. 2025 Aug 2;41(8):btaf417. doi: 10.1093/bioinformatics/btaf417. Bioinformatics. 2025. PMID: 40728934 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

