A telomere-to-telomere gapless genome reveals SlPRR1 control of circadian rhythm and photoperiodic flowering in tomato
- PMID: 40601421
- PMCID: PMC12218202
- DOI: 10.1093/gigascience/giaf058
A telomere-to-telomere gapless genome reveals SlPRR1 control of circadian rhythm and photoperiodic flowering in tomato
Abstract
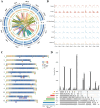
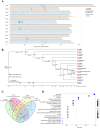
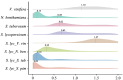
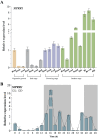
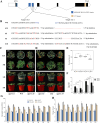
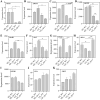
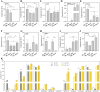
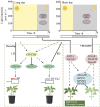
Cultivated tomato (Solanum lycopersicum) is a major vegetable crop of high economic value that serves as an important model for studying flowering time in day-neutral plants. A complete, continuous, and gapless genome of cultivated tomato is essential for genetic research and breeding programs. Here, we report the construction of a telomere-to-telomere (T2T) gap-free genome of S. lycopersicum cv. VF36 using a combination of sequencing technologies. The 815.27-Mb T2T "VF36" genome contained 600.23 Mb of transposable elements. Through comparative genomics and phylogenetic analysis, we identified structural variations between the "VF36" and "Heinz 1706" genomes and found no evidence of a recent species-specific whole-genome duplication in the "VF36" tomato. Furthermore, a core circadian oscillator, SlPRR1, was identified, which peaked at night in a circadian rhythm. CRISPR/Cas9-mediated knockdown of SlPRR1 in tomatoes demonstrated that slprr1 mutant lines exhibited significantly earlier flowering under long-day condition than wild type. We present a hypothetical model of how SlPRR1 regulates flowering time and chlorophyll biosynthesis in response to photoperiod. This T2T genomic resource will accelerate the genetic improvement of large-fruited tomatoes, and the SlPRR1-related hypothetical model will enhance our understanding of the photoperiodic response in cultivated tomatoes, revealing a regulatory mechanism for manipulating flowering time.
Keywords: SlPRR1; chlorophyll biosynthesis; cultivated tomato T2T genome; flowering time; photoperiod.
© The Author(s) 2025. Published by Oxford University Press GigaScience.
Conflict of interest statement
The authors declare that they have no competing interests.
Figures

Similar articles
-
Gene editing of clock components in Solanum lycopersicum: Effects on gene expression, development, and productivity.Plant J. 2025 Jul;123(2):e70383. doi: 10.1111/tpj.70383. Plant J. 2025. PMID: 40726316
-
Gap-free genome and efficient transcript purification system reveals the genomes diversity and chlorophyll degradation mechanism in pitaya.J Integr Plant Biol. 2025 Jul;67(7):1771-1786. doi: 10.1111/jipb.13925. Epub 2025 Jun 5. J Integr Plant Biol. 2025. PMID: 40471007
-
A telomere-to-telomere gap-free assembly integrating multi-omics uncovers the genetic mechanism of fruit quality and important agronomic trait associations in pomegranate.Plant Biotechnol J. 2025 Jul;23(7):2852-2870. doi: 10.1111/pbi.70107. Epub 2025 May 3. Plant Biotechnol J. 2025. PMID: 40318230 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2017 Dec 22;12(12):CD011535. doi: 10.1002/14651858.CD011535.pub2. Cochrane Database Syst Rev. 2017. Update in: Cochrane Database Syst Rev. 2020 Jan 9;1:CD011535. doi: 10.1002/14651858.CD011535.pub3. PMID: 29271481 Free PMC article. Updated.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

