Proteome remodeling in the zoospore-to-vegetative cell transition of the stramenopile Aurantiochytrium limacinum reveals candidate ectoplasmic network proteins
- PMID: 40601697
- PMCID: PMC12221091
- DOI: 10.1371/journal.pone.0326651
Proteome remodeling in the zoospore-to-vegetative cell transition of the stramenopile Aurantiochytrium limacinum reveals candidate ectoplasmic network proteins
Abstract
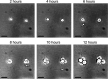
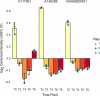
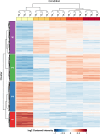
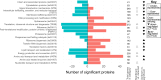
Thraustochytrids are marine protists of ecological and biotechnological importance. Like many other eukaryotes, their life cycle includes a critical transition from a flagellated, swimming zoospore dispersal stage to a settled, surface-attached, growing vegetative cell. Unlike other eukaryotes, the settling vegetative cells of thraustochytrids (and their labyrinthulomycete relatives) attach to surfaces by producing a unique structure known as the ectoplasmic network, and its associated connection to the cytoplasm, the bothrosome. We conducted time-course proteomics and microscopy to study this transition in the model thraustochytrid Aurantiochytrium limacinum ATCC MYA-1381. We identified 623 proteins significantly differentially expressed between zoospores and samples collected 2, 4, 6, and 8 hours after settlement. Analysis of the differentially expressed proteins revealed broad cellular changes during the transition from zoospore to vegetative cell, including shifts in motility, signaling, and metabolism. A relative enrichment of proteasomal and ribosomal components in the zoospores suggests these proteins are stockpiled, priming the zoospore for rapid protein turnover upon settlement. Flagellar proteins were strongly downregulated upon settlement, coinciding with loss of motility. Environmental sensing systems, such as channelrhodopsins, declined post-settlement. The proteomic changes also suggest that zoospores rely on catabolism of stored lipids by beta-oxidation, whereas settled vegetative cells shift towards anabolic metabolism, including gluconeogenesis (growth media contained glycerol), and the biosynthesis of membrane lipids, amino acids, and nucleic acids. A search for proteins which were upregulated during vegetative cell settlement, and which were phylogenetically divergent in thraustochytrids, yielded a list of potential ectoplasmic network or bothrosome candidates, including potential homologs of micronemal adhesins and membrane-trafficking proteins. Our findings illuminate a critical life-history transition in A. limacinum, and identify targets for understanding the evolutionary origins and functions of unique labyrinthulomycete structures.
Copyright: © 2025 Gil-Gomez et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures

References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

