Single-cell analysis of the decidua unveils the mechanism of anti-inflammatory exosomes for chorioamnionitis in nonhuman primates
- PMID: 40601753
- PMCID: PMC12219548
- DOI: 10.1126/sciadv.adp0467
Single-cell analysis of the decidua unveils the mechanism of anti-inflammatory exosomes for chorioamnionitis in nonhuman primates
Abstract
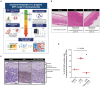
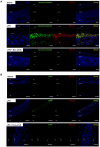
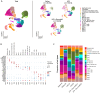
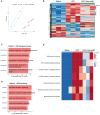
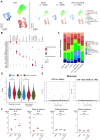
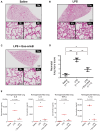
The effectiveness of exosomes engineered to carry a dominantly active variant of inhibitor α of nuclear factor κB (NF-κB) (IκBα), super-repressor IκB (srIκB), that inhibits the expression of NF-κB in various animal models of inflammatory diseases has been demonstrated. In this study, we used a lipopolysaccharide (LPS)-induced chorioamnionitis model in pregnant nonhuman primates to explore the therapeutic potential and mode of action of srIκB-loaded exosomes (Exo-srIκBs). Intraamniotic injection of LPS induced infiltration of BCL2A1-positive neutrophils and CD68-positive macrophages in the extraplacental membranes, causing fetal lung injury. Conversely, administration of Exo-srIκB via intraamniotic and intravenous routes (6.9 × 1010 and 4 × 1011 particle numbers, respectively) ameliorated these effects. Single-cell RNA sequencing of the decidua and bulk RNA sequencing of the choriodecidua highlighted that Exo-srIκB treatment mitigated LPS-induced inflammatory pathways, particularly in macrophages, leading to a cascade effect on neutrophils through NF-κB signaling inhibition. These findings underscore the potential of Exo-srIκB as a therapeutic strategy for chorioamnionitis.
Figures


References
-
- J. R. Fowler, L. V. Simon, “Chorioamnionitis,” in StatPearls, NBK532251 (Treasure Island, 2023).
-
- Perkins R. P., Zhou S. M., Butler C., Skipper B. J., Histologic chorioamnionitis in pregnancies of various gestational ages: Implications in preterm rupture of membranes. Obstet. Gynecol. 70, 856–860 (1987). - PubMed
-
- Romero R., Mazor M., Infection and preterm labor. Clin. Obstet. Gynecol. 31, 553–584 (1988). - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

