A genome-wide in vivo CRISPR activation screen identifies BACE1 as a therapeutic vulnerability of lung cancer brain metastasis
- PMID: 40601773
- PMCID: PMC7617989
- DOI: 10.1126/scitranslmed.adu2459
A genome-wide in vivo CRISPR activation screen identifies BACE1 as a therapeutic vulnerability of lung cancer brain metastasis
Abstract
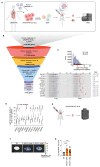
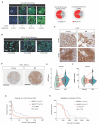
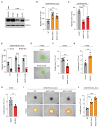
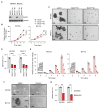
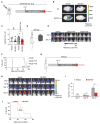
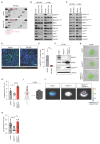
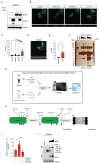
Brain metastasis occurs in up to 40% of patients with non-small cell lung cancer (NSCLC). Considerable genomic heterogeneity exists between the primary lung tumor and respective brain metastasis; however, the identity of the genes capable of driving brain metastasis is incompletely understood. Here, we carried out an in vivo genome-wide CRISPR activation screen to identify molecular drivers of brain metastasis from an orthotopic xenograft model derived from a patient with NSCLC. We found that activating expression of the Alzheimer's disease-associated beta-secretase 1 (BACE1) led to a substantial increase in brain metastases. Furthermore, genetic and pharmacological inhibition of BACE1 blocked NSCLC brain metastasis. Mechanistically, we identified that BACE1 acts through epidermal growth factor receptor to drive this metastatic phenotype. Together, our data highlight the power of in vivo CRISPR activation screening to unveil molecular drivers and potential therapeutic targets of NSCLC brain metastasis.
Conflict of interest statement
S.B. and K.Z are listed as inventors in an issued patent application (US patent# 11559528) related to this study. Other authors declare no competing interests relevant to the current study.
Figures
References
-
- Aizer AA, Lamba N, Ahluwalia MS, Aldape K, Boire A, Brastianos PK, Brown PD, Camidge DR, Chiang VL, Davies MA, Hu LS, et al. Brain metastases: A Society for Neuro-Oncology (SNO) consensus review on current management and future directions. Neuro Oncol. 2022;24:1613–1646. doi: 10.1093/neuonc/noac118. - DOI - PMC - PubMed
-
- Achrol AS, Rennert RC, Anders C, Soffietti R, Ahluwalia MS, Nayak L, Peters S, Arvold ND, Harsh GR, Steeg PS, Chang SD. Brain metastases. Nat Rev Dis Primers. 2019;5:5. - PubMed
-
- Cagney DN, Martin AM, Catalano PJ, Redig AJ, Lin NU, Lee EQ, Wen PY, Dunn IF, Bi WL, Weiss SE, Haas-Kogan DA, et al. Incidence and prognosis of patients with brain metastases at diagnosis of systemic malignancy: a population-based study. Neuro Oncol. 2017;19:1511–1521. doi: 10.1093/neuonc/nox077. - DOI - PMC - PubMed
-
- Janne PA, Riely GJ, Gadgeel SM, Heist RS, Ou SI, Pacheco JM, Johnson ML, Sabari JK, Leventakos K, Yau E, Bazhenova L, et al. Adagrasib in Non-Small-Cell Lung Cancer Harboring a KRAS(G12C) Mutation. N Engl J Med. 2022;387:120–131. - PubMed
-
- Soria JC, Ohe Y, Vansteenkiste J, Reungwetwattana T, Chewaskulyong B, Lee KH, Dechaphunkul A, Imamura F, Nogami N, Kurata T, Okamoto I, et al. Osimertinib in Untreated EGFR-Mutated Advanced Non-Small-Cell Lung Cancer. N Engl J Med. 2018;378:113–125. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

