Differential Efficacy of Bevacizumab and Erlotinib in Preclinical Models of Renal Medullary Carcinoma and Fumarate Hydratase-Deficient Renal Cell Carcinoma
- PMID: 40601845
- PMCID: PMC12323885
- DOI: 10.1158/1535-7163.MCT-24-0703
Differential Efficacy of Bevacizumab and Erlotinib in Preclinical Models of Renal Medullary Carcinoma and Fumarate Hydratase-Deficient Renal Cell Carcinoma
Abstract
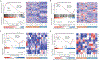
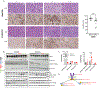
Renal medullary carcinoma (RMC) and fumarate hydratase (FH)-deficient renal cell carcinoma (RCC) are rare and highly aggressive cancers. Although the combination of vascular endothelial growth factor (VEGF) inhibition by bevacizumab and epidermal growth factor receptor (EGFR) inhibition by erlotinib is clinically used for both diseases, the differential effect of each component has not been investigated. Transcriptomic profiling revealed that RMC and FH-deficient tumor tissues demonstrate increased EGFR but not VEGF expression compared with adjacent normal kidney. Subsequent in vitro studies revealed that RMC and FH-deficient cell lines are sensitive to erlotinib treatment, whereas clear cell RCC cell lines are resistant. We developed patient-derived xenograft (PDX) models of tumors exposed to first-line therapies to represent treatment-experienced RMC and FH-deficient RCC models. These models were then used to determine tumor growth response to angiogenesis inhibition by bevacizumab alone or in combination with erlotinib. The FH-deficient RCC PDX model responded to either bevacizumab or erlotinib alone or in combination while the RMC PDX responded only to erlotinib consistent with clinical and preclinical data suggesting that RMC is refractory to angiogenesis inhibition. Statistically higher expression of EGFR was observed in the RMC PDX model compared to the FH-deficient model; while higher phosphorylated tyrosine-416 SRC expression was observed in FH-deficient PDX model compared to the RMC model. Our preclinical data suggest that EGFR signaling differentially modulates tumor growth in RMC and FH-deficient RCC and that angiogenesis inhibition is a valid target in FH-deficient RCC but not RMC.
Conflict of interest statement
Pavlos Msaouel has received honoraria for service on a Scientific Advisory Board for Mirati Therapeutics, Bristol Myers Squibb, and Exelixis; consulting for Axiom Healthcare Strategies; non-branded educational programs supported by DAVA Oncology, Exelixis and Pfizer; and research funding for clinical trials from Takeda, Bristol Myers Squibb, Mirati Therapeutics, Gateway for Cancer Research, and the University of Texas MD Anderson Cancer Center. Jose Karam has acted as a consultant, served on the advisory board, or has accepted an honorarium from Pfizer, Merck, Johnson and Johnson, Syapse, American Board of Urology, American Urological Association, Kidney Cancer Association, Texas Urological Society, Clarivate, Specialty Networks/MJH, Medscape, PCORI, and Guidepoint Global; he has received research funding from Roche/Genetech, Mirati, Merck and Elypta; and he owns stock in MedTek and ROM Technologies.
Figures


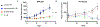
Similar articles
-
A Phase 2 Trial of Talazoparib and Avelumab in Genomically Defined Metastatic Kidney Cancer.Eur Urol Oncol. 2024 Aug;7(4):804-811. doi: 10.1016/j.euo.2023.10.017. Epub 2023 Nov 7. Eur Urol Oncol. 2024. PMID: 37945488 Free PMC article. Clinical Trial.
-
The impact of the new WHO classification of renal cell carcinoma on the diagnosis of hereditary leiomyomatosis and renal cell carcinoma.Nephrol Dial Transplant. 2025 Jun 30;40(7):1428-1432. doi: 10.1093/ndt/gfaf032. Nephrol Dial Transplant. 2025. PMID: 39979023 Free PMC article. Review.
-
Targeting NAD+ Metabolism Vulnerability in FH-Deficient Hereditary Leiomyomatosis and Renal Cell Carcinoma with the Novel NAMPT Inhibitor OT-82.Mol Cancer Ther. 2025 Feb 4;24(2):200-213. doi: 10.1158/1535-7163.MCT-24-0225. Mol Cancer Ther. 2025. PMID: 39397296 Free PMC article.
-
Dual Inhibition of CDK4/6 and mTORC1 Establishes a Preclinical Strategy for Translocation Renal Cell Carcinoma.bioRxiv [Preprint]. 2025 Jul 17:2025.07.11.663903. doi: 10.1101/2025.07.11.663903. bioRxiv. 2025. PMID: 40747423 Free PMC article. Preprint.
-
First-line treatment of advanced epidermal growth factor receptor (EGFR) mutation positive non-squamous non-small cell lung cancer.Cochrane Database Syst Rev. 2016 May 25;(5):CD010383. doi: 10.1002/14651858.CD010383.pub2. Cochrane Database Syst Rev. 2016. Update in: Cochrane Database Syst Rev. 2021 Mar 18;3:CD010383. doi: 10.1002/14651858.CD010383.pub3. PMID: 27223332 Updated.
References
-
- Skala SL, Dhanasekaran SM, Mehra R Hereditary Leiomyomatosis and Renal Cell Carcinoma Syndrome (HLRCC): A Contemporary Review and Practical Discussion of the Differential Diagnosis for HLRCC-Associated Renal Cell Carcinoma. Arch Pathol Lab Med 2018;142:1202–1215. - PubMed
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

