Cardiolipin-mimic lipid nanoparticles without antibody modification delivered senolytic in vivo CAR-T therapy for inflamm-aging
- PMID: 40602406
- PMCID: PMC12281384
- DOI: 10.1016/j.xcrm.2025.102209
Cardiolipin-mimic lipid nanoparticles without antibody modification delivered senolytic in vivo CAR-T therapy for inflamm-aging
Abstract
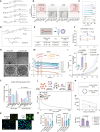
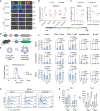
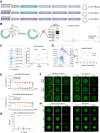
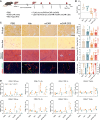
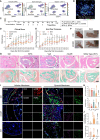
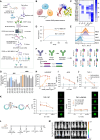
mRNA-based in vivo chimeric antigen receptor (CAR)-T cell engineering offers advantages over ex vivo therapies, including streamlined manufacturing and transient expression. However, current delivery methods require antibody-modified vehicles with manufacturing challenges. In this study, inspired by cardiolipin, we identify cardiolipin-like di-phosphoramide lipids that improve T cell transfection without targeting ligands, both in vitro and in vivo. The T cell-favored tropism is likely due to the lipid's packing, shape, and rigidity. Encapsulating circular RNA further prolongs mRNA expression in the spleen and T cells. Using PL40 lipid nanoparticles, we deliver mRNA encoding a CAR targeting the senolytic and inflammatory antigen urokinase-type plasminogen activator receptor (uPAR), alleviating uPAR-related liver fibrosis and rheumatoid arthritis (RA). Single-cell sequencing in humans confirms uPAR's relevance to senescence and inflammation in RA. To facilitate clinical translation, we screen and humanize single-chain variable fragments (scFvs) against uPAR, establishing a PL40 mRNA-encoded humanized uPAR CAR with potential for treating aging-inflamed disorders.
Keywords: T cells; aging-inflamed disorders; chimeric antigen receptor: CAR; circular mRNA; senolytic; uPAR.
Copyright © 2025 The Authors. Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests L.M., B.M., and Z.Z. have filed a patent for the development of CAMP lipids. M.D., L.M., X.G., Z.Z., and Z.L. have filed a patent for the human uPAR targeting CARs and antibodies.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

