Overcoming multidrug resistance in gastrointestinal cancers with a CDH17-targeted ADC conjugated to a DNA topoisomerase inhibitor
- PMID: 40602407
- PMCID: PMC12281427
- DOI: 10.1016/j.xcrm.2025.102213
Overcoming multidrug resistance in gastrointestinal cancers with a CDH17-targeted ADC conjugated to a DNA topoisomerase inhibitor
Abstract
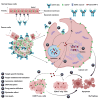
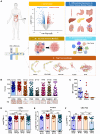
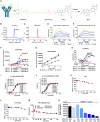
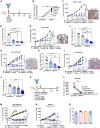
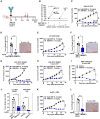
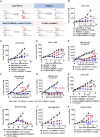
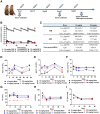
Cadherin 17 (CDH17) has emerged as a promising target for gastrointestinal (GI) cancers, which are often complicated by multidrug resistance (MDR) and recurrence. In this study, we developed 7MW4911, a CDH17-targeted antibody-drug conjugate (ADC) that incorporates a topoisomerase inhibitor MF-6 (Topi MF-6) payload linked via a cleavable linker, designed specifically to address MDR in GI cancers. 7MW4911 exhibited high specificity for CDH17-expressing cancer cells and potent cytotoxicity in vitro. In preclinical models, including patient-derived xenografts (PDXs) with distinct mutations, 7MW4911 achieved tumor growth inhibition ranging from 71% to 99%. Remarkably, 7MW4911 outperformed monomethyl auristatin E (MMAE)-based and Deruxtecan (DXd)-based ADCs in MDR models, highlighting its effectiveness against drug-resistant cancer phenotypes. Additionally, 7MW4911 showed favorable pharmacokinetics and a highest non-severely toxic dose (HNSTD) exceeding 20 mg/kg in cynomolgus monkeys, underscoring its promising safety profile. Together, these findings position 7MW4911 as a promising ADC candidate capable of enhancing therapeutic outcomes in GI cancers.
Keywords: ADC; CDH17; colorectal cancer; gastrointestinal cancers; multidrug resistance.
Copyright © 2025 The Author(s). Published by Elsevier Inc. All rights reserved.
Conflict of interest statement
Declaration of interests All authors are employees of Mabwell (Shanghai) Bioscience Co., Ltd., or Jiangsu Mabwell Health Pharmaceutical R&D Co., Ltd., and may hold shares in Mabwell (Shanghai) Bioscience Co., Ltd. Rui Wang, P.F., W.Z., X.T., X.G., and D.L. are listed as inventors on a patent application for the anti-CDH17 ADC 7MW4911.
Figures
References
-
- Baraniskin A., Van Laethem J.-L., Wyrwicz L., Guller U., Wasan H.S., Matysiak-Budnik T., Gruenberger T., Ducreux M., Carneiro F., Van Cutsem E., et al. Clinical relevance of molecular diagnostics in gastrointestinal (GI) cancer: European Society of Digestive Oncology (ESDO) expert discussion and recommendations from the 17th European Society for Medical Oncology (ESMO)/World Congress on Gastrointestinal Cancer, Barcelona. Eur. J. Cancer. 2017;86:305–317. doi: 10.1016/j.ejca.2017.09.021. - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

