CalTrig: A GUI-Based Machine Learning Approach for Decoding Neuronal Calcium Transients in Freely Moving Rodents
- PMID: 40603011
- PMCID: PMC12263099
- DOI: 10.1523/ENEURO.0009-25.2025
CalTrig: A GUI-Based Machine Learning Approach for Decoding Neuronal Calcium Transients in Freely Moving Rodents
Abstract
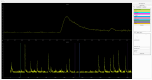
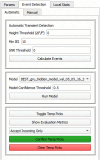
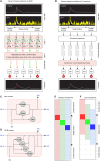
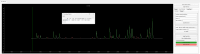
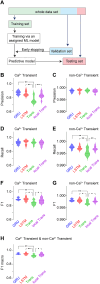
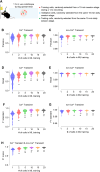
Advances in in vivo Ca2+ imaging using miniature microscopes have enabled researchers to study single-neuron activity in freely moving animals. Tools such as Minian and CalmAn have been developed to convert Ca2+ visual signals to numerical data, collectively referred to as CalV2N. However, substantial challenges remain in analyzing the large datasets generated by CalV2N, particularly in integrating data streams, evaluating CalV2N output quality, and reliably and efficiently identifying Ca2+ transients. In this study, we introduce CalTrig, an open-source graphical user interface (GUI) tool designed to address these challenges at the post-CalV2N stage of data processing collected from C57BL/6J mice. CalTrig integrates multiple data streams, including Ca2+ imaging, neuronal footprints, Ca2+ traces, and behavioral tracking, and offers capabilities for evaluating the quality of CalV2N outputs. It enables synchronized visualization and efficient Ca2+ transient identification. We evaluated four machine learning models (i.e., GRU, LSTM, Transformer, and Local Transformer) for Ca2+ transient detection. Our results indicate that the GRU model offers the highest predictability and computational efficiency, achieving stable performance across training sessions, different animals, and even among different brain regions. The integration of manual, parameter-based, and machine learning-based detection methods in CalTrig provides flexibility and accuracy for various research applications. The user-friendly interface and low computing demands of CalTrig make it accessible to neuroscientists without programming expertise. We further conclude that CalTrig enables deeper exploration of brain function, supports hypothesis generation about neuronal mechanisms, and opens new avenues for understanding neurological disorders and developing treatments.
Keywords: GRU; calcium transients; data visualization; in vivo calcium imaging; machine learning; miniScope.
Copyright © 2025 Lange et al.
Conflict of interest statement
The authors declare no competing financial interests.
Figures





Update of
-
CalTrig: A GUI-based Machine Learning Approach for Decoding Neuronal Calcium Transients in Freely Moving Rodents.bioRxiv [Preprint]. 2024 Nov 19:2024.09.30.615860. doi: 10.1101/2024.09.30.615860. bioRxiv. 2024. Update in: eNeuro. 2025 Jul 15;12(7):ENEURO.0009-25.2025. doi: 10.1523/ENEURO.0009-25.2025. PMID: 39372793 Free PMC article. Updated. Preprint.
Similar articles
-
CalTrig: A GUI-based Machine Learning Approach for Decoding Neuronal Calcium Transients in Freely Moving Rodents.bioRxiv [Preprint]. 2024 Nov 19:2024.09.30.615860. doi: 10.1101/2024.09.30.615860. bioRxiv. 2024. Update in: eNeuro. 2025 Jul 15;12(7):ENEURO.0009-25.2025. doi: 10.1523/ENEURO.0009-25.2025. PMID: 39372793 Free PMC article. Updated. Preprint.
-
Monitoring Adverse Drug Events in Web Forums: Evaluation of a Pipeline and Use Case Study.J Med Internet Res. 2024 Jun 18;26:e46176. doi: 10.2196/46176. J Med Internet Res. 2024. PMID: 38888956 Free PMC article.
-
Comparison of Two Modern Survival Prediction Tools, SORG-MLA and METSSS, in Patients With Symptomatic Long-bone Metastases Who Underwent Local Treatment With Surgery Followed by Radiotherapy and With Radiotherapy Alone.Clin Orthop Relat Res. 2024 Dec 1;482(12):2193-2208. doi: 10.1097/CORR.0000000000003185. Epub 2024 Jul 23. Clin Orthop Relat Res. 2024. PMID: 39051924
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Generalizable machine learning for stress monitoring from wearable devices: A systematic literature review.Int J Med Inform. 2023 May;173:105026. doi: 10.1016/j.ijmedinf.2023.105026. Epub 2023 Feb 28. Int J Med Inform. 2023. PMID: 36893657
Cited by
-
Decoding Secondary Motor Cortex Neuronal Activity During Cocaine Self-Administration: Insights From Longitudinal In Vivo Calcium Imaging.Biol Psychiatry Glob Open Sci. 2025 May 9;5(5):100531. doi: 10.1016/j.bpsgos.2025.100531. eCollection 2025 Sep. Biol Psychiatry Glob Open Sci. 2025. PMID: 40703665 Free PMC article.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous
