Ensemble methods and partially-supervised learning for accurate and robust automatic murine organ segmentation
- PMID: 40603458
- PMCID: PMC12222911
- DOI: 10.1038/s41598-025-05954-6
Ensemble methods and partially-supervised learning for accurate and robust automatic murine organ segmentation
Abstract
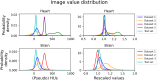
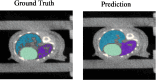
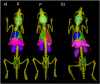
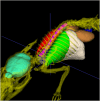
Delineation of multiple organs in murine µCT images is crucial for preclinical studies but requires manual volumetric segmentation, a tedious and time-consuming process prone to inter-observer variability. Automatic deep learning-based segmentation can improve speed and reproducibility. While 2D and 3D deep learning models have been developed for anatomical segmentation, their generalization to external datasets has not been extensively investigated. Furthermore, ensemble learning, combining predictions of multiple 2D models, and partially-supervised learning (PSL), enabling training on partially-labeled datasets, have not been explored for preclinical purposes. This study demonstrates the first use of PSL frameworks and the superiority of 3D models in accuracy and generalizability to external datasets. Ensemble methods performed on par or better than the best individual 2D network, but only 3D models consistently generalized to external datasets (Dice Similarity Coefficient (DSC) > 0.8). PSL frameworks showed promising results across various datasets and organs, but its generalization to external data can be improved for some organs. This work highlights the superiority of 3D models over 2D and ensemble counterparts in accuracy and generalizability for murine µCT image segmentation. Additionally, a promising PSL framework is presented for leveraging multiple datasets without complete annotations. Our model can increase time-efficiency and improve reproducibility in preclinical radiotherapy workflows by circumventing manual contouring bottlenecks. Moreover, high segmentation accuracy of 3D models allows monitoring multiple organs over time using repeated µCT imaging, potentially reducing the number of mice sacrificed in studies, adhering to the 3R principle, specifically Reduction and Refinement.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Semi-Supervised Learning Allows for Improved Segmentation With Reduced Annotations of Brain Metastases Using Multicenter MRI Data.J Magn Reson Imaging. 2025 Jun;61(6):2469-2479. doi: 10.1002/jmri.29686. Epub 2025 Jan 10. J Magn Reson Imaging. 2025. PMID: 39792624 Free PMC article.
-
A preprocessing method based on 3D U-Net for abdomen segmentation.Comput Biol Med. 2025 Sep;196(Pt A):110709. doi: 10.1016/j.compbiomed.2025.110709. Epub 2025 Jul 12. Comput Biol Med. 2025. PMID: 40652756
-
Automatic Segmentation and Alignment of Uterine Shapes from 3D Ultrasound Data.Comput Biol Med. 2024 Aug;178:108794. doi: 10.1016/j.compbiomed.2024.108794. Epub 2024 Jun 27. Comput Biol Med. 2024. PMID: 38941903
-
Transformers for Neuroimage Segmentation: Scoping Review.J Med Internet Res. 2025 Jan 29;27:e57723. doi: 10.2196/57723. J Med Internet Res. 2025. PMID: 39879621 Free PMC article.
-
Deep learning algorithm performance in contouring head and neck organs at risk: a systematic review and single-arm meta-analysis.Biomed Eng Online. 2023 Nov 1;22(1):104. doi: 10.1186/s12938-023-01159-y. Biomed Eng Online. 2023. PMID: 37915046 Free PMC article.
References
-
- Verhaegen, F., Granton, P. & Tryggestad, E. Small animal radiotherapy research platforms. Phys. Med. Biol.56, R55–R83. 10.1088/0031-9155/56/12/R01 (2011). - PubMed
MeSH terms
LinkOut - more resources
Full Text Sources

