FAM72A promotes UNG2 degradation and mutagenesis in human cancer cells
- PMID: 40604025
- PMCID: PMC12223117
- DOI: 10.1038/s41598-025-07723-x
FAM72A promotes UNG2 degradation and mutagenesis in human cancer cells
Abstract
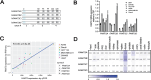
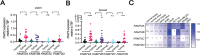
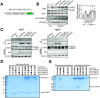
Genetic lesions drive cancer development and progression, and understanding their origins will reveal the mechanisms of carcinogenesis. We showed that murine FAM72A promotes mutagenic DNA repair during antibody maturation by acting as a substrate adaptor of the CTLHMKLN1 E3 ligase to induce the proteasome degradation of Uracil DNA glycosylase 2 (UNG2), a pivotal enzyme of the base excision repair. In humans, the FAM72 gene has expanded to include four paralogues named FAM72A-D. Bioinformatic studies suggested that the human FAM72 genes are overexpressed in a broad range of cancers. However, the functional roles of FAM72A-D in human biology and cancer are unknown. Here, we show that FAM72 family members are minimally expressed in most healthy tissues except for thymus, and that FAM72A, B and D are overexpressed in primary tumorigenic tissues. Human FAM72 expression inversely correlates with UNG2 protein level in human cell lines and primary tumorigenic tissues suggesting that human FAM72 promotes UNG2 degradation. However, only FAM72A is able to bind to and induce UNG2 degradation in human cells. Our results suggest that the ability of FAM72A to induce UNG2 degradation contributes to neoplasia in a variety of cancer types by promoting mutagenic repair of genomic dUs.
Keywords: Base excision repair; Cancer; Cytosine deamination; FAM72; Mutation; Uracil DNA glycosylase 2.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Competing interests: The authors declare no competing interests. Conflict of interest: F.S is a founder and consultant for Repare Therapeutics and Induxion Therapeutics.
Figures

Similar articles
-
FAM72A degrades UNG2 through the GID/CTLH complex to promote mutagenic repair during antibody maturation.Nat Commun. 2024 Aug 30;15(1):7541. doi: 10.1038/s41467-024-52009-x. Nat Commun. 2024. PMID: 39215025 Free PMC article.
-
MKLN1-dependent GID4/CTLH E3 ubiquitin ligase complex assemblies are required to support B-cell antibody diversification.J Immunol. 2025 Aug 20:vkaf201. doi: 10.1093/jimmun/vkaf201. Online ahead of print. J Immunol. 2025. PMID: 40838616
-
FAM72A antagonizes UNG2 to promote mutagenic repair during antibody maturation.Nature. 2021 Dec;600(7888):324-328. doi: 10.1038/s41586-021-04144-4. Epub 2021 Nov 24. Nature. 2021. PMID: 34819670 Free PMC article.
-
Role of NEIL1 in genome maintenance.DNA Repair (Amst). 2025 Apr;148:103820. doi: 10.1016/j.dnarep.2025.103820. Epub 2025 Feb 19. DNA Repair (Amst). 2025. PMID: 40010204 Review.
-
Impact of DNA polymorphisms in key DNA base excision repair proteins on cancer risk.Hum Exp Toxicol. 2012 Oct;31(10):981-1005. doi: 10.1177/0960327112444476. Epub 2012 Sep 27. Hum Exp Toxicol. 2012. PMID: 23023028 Free PMC article.
References
-
- Methot, S. P. & Di Noia, J. M. Molecular mechanisms of somatic hypermutation and class switch recombination. Adv. Immunol.133, 37–87. 10.1016/bs.ai.2016.11.002 (2017). - PubMed
-
- Cascalho, M., Wong, J., Steinberg, C. & Wabl, M. Mismatch repair co-opted by hypermutation. Science279, 1207–1210 (1998). - PubMed
-
- Martin, A. & Scharff, M. D. AID and mismatch repair in antibody diversification. Nat. Rev. Immunol.2, 605–614 (2002). - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical

