PPP2R1A mutations portend improved survival after cancer immunotherapy
- PMID: 40604275
- PMCID: PMC12350166
- DOI: 10.1038/s41586-025-09203-8
PPP2R1A mutations portend improved survival after cancer immunotherapy
Erratum in
-
Author Correction: PPP2R1A mutations portend improved survival after cancer immunotherapy.Nature. 2025 Sep;645(8082):E10. doi: 10.1038/s41586-025-09586-8. Nature. 2025. PMID: 40913056 Free PMC article. No abstract available.
Abstract
Immune checkpoint blockade (ICB) therapy is effective against many cancers, although resistance remains a major issue and new strategies are needed to improve clinical outcomes1-5. Here we studied ICB response in a cohort of patients with ovarian clear cell carcinoma-a cancer type that poses considerable clinical challenges and lacks effective therapies6-8. We observed significantly prolonged overall survival and progression-free survival in patients with tumours with PPP2R1A mutations. Importantly, our findings were validated in additional ICB-treated patient cohorts across multiple cancer types. Translational analyses from tumour biopsies demonstrated enhanced IFNγ signalling, and the presence of tertiary lymphoid structures at the baseline, as well as enhanced immune infiltration and expansion of CD45RO+CD8+ T cells in the tumour neighbourhood after ICB treatment in PPP2R1A-mutated tumours. Parallel preclinical investigations showed that targeting PPP2R1A (by pharmacological inhibition or genetic modifications) in in vitro and in vivo models was associated with improved survival in the setting of treatment with several forms of immunotherapy, including chimeric antigen receptor (CAR)-T cell therapy and ICB. The results from these studies suggest that therapeutic targeting of PPP2R1A may represent an effective strategy to improve patient outcomes after ICB or other forms of immunotherapy, although additional mechanistic and therapeutic insights are needed.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: A.K.S. reports a consulting role at Merck, Onxeo, ImmunoGen, GSK, Astra Zeneca and Iylon. The other authors declare no competing interests.
Figures


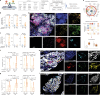


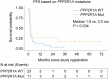

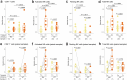





References
-
- Matulonis, U. A. et al. Antitumor activity and safety of pembrolizumab in patients with advanced recurrent ovarian cancer: results from the phase II KEYNOTE-100 study. Ann. Oncol.30, 1080–1087 (2019). - PubMed
MeSH terms
Substances
Grants and funding
- T32 CA101642/CA/NCI NIH HHS/United States
- P30 CA016672/CA/NCI NIH HHS/United States
- R01 CA276569/CA/NCI NIH HHS/United States
- K07 CA201013/CA/NCI NIH HHS/United States
- P30 CA010815/CA/NCI NIH HHS/United States
- R01 CA239128/CA/NCI NIH HHS/United States
- M01 RR000847/RR/NCRR NIH HHS/United States
- R01 CA260661/CA/NCI NIH HHS/United States
- P50 CA272218/CA/NCI NIH HHS/United States
- P50 CA281701/CA/NCI NIH HHS/United States
- L30 CA123720/CA/NCI NIH HHS/United States
- R01 CA202919/CA/NCI NIH HHS/United States
- S10 OD024977/OD/NIH HHS/United States
- R01 CA243142/CA/NCI NIH HHS/United States
- R01 CA266280/CA/NCI NIH HHS/United States
- R01 CA306089/CA/NCI NIH HHS/United States
- U01 CA294518/CA/NCI NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

