Mapping and engineering RNA-driven architecture of the multiphase nucleolus
- PMID: 40604277
- PMCID: PMC12350172
- DOI: 10.1038/s41586-025-09207-4
Mapping and engineering RNA-driven architecture of the multiphase nucleolus
Abstract
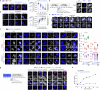
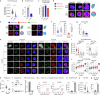
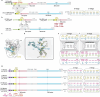
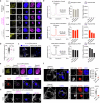
Biomolecular condensates are key features of intracellular compartmentalization1,2. As the most prominent nuclear condensate in eukaryotes, the nucleolus is a multiphase liquid-like structure in which ribosomal RNAs (rRNAs) are transcribed and processed, undergoing multiple maturation steps to form the small (SSU) and large (LSU) ribosomal subunits3-5. However, how rRNA processing is coupled to the layered organization of the nucleolus is poorly understood owing to a lack of tools to precisely monitor and perturb nucleolar rRNA processing dynamics. Here we developed two complementary approaches to spatiotemporally map rRNA processing and engineer de novo nucleoli. Using sequencing in parallel with imaging, we found that rRNA processing steps are spatially segregated, with sequential maturation of rRNA required for its outward movement through nucleolar phases. By generating synthetic nucleoli in cells using an engineered rDNA plasmid system, we show that defects in SSU processing can alter the ordering of nucleolar phases, resulting in inside-out nucleoli and preventing rRNA outflux, while LSU precursors are necessary to build the outermost layer of the nucleolus. These findings demonstrate how rRNA is both a scaffold and substrate for the nucleolus, with rRNA acting as a programmable blueprint for the multiphase architecture that facilitates assembly of an essential molecular machine.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: C.P.B. is a scientific founder, scientific advisory board member, shareholder and consultant for Nereid Therapeutics. The other authors declare no competing interests.
Figures






Update of
-
Mapping and engineering RNA-controlled architecture of the multiphase nucleolus.bioRxiv [Preprint]. 2024 Sep 29:2024.09.28.615444. doi: 10.1101/2024.09.28.615444. bioRxiv. 2024. Update in: Nature. 2025 Aug;644(8076):557-566. doi: 10.1038/s41586-025-09207-4. PMID: 39386460 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

