Rewiring endogenous genes in CAR T cells for tumour-restricted payload delivery
- PMID: 40604285
- PMCID: PMC12328239
- DOI: 10.1038/s41586-025-09212-7
Rewiring endogenous genes in CAR T cells for tumour-restricted payload delivery
Abstract
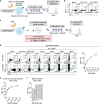
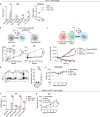
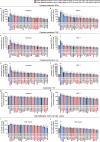
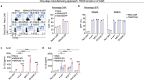
The efficacy of chimeric antigen receptor (CAR) T cell therapy in solid tumours is limited by immunosuppression and antigen heterogeneity1-3. To overcome these barriers, 'armoured' CAR T cells, which secrete proinflammatory cytokines, have been developed4. However, their clinical application has been limited because of toxicity related to peripheral expression of the armouring transgene5. Here, we have developed a CRISPR knock-in strategy that leverages the regulatory mechanisms of endogenous genes to drive transgene expression in a tumour-localized manner. By screening endogenous genes with tumour-restricted expression, we have identified the NR4A2 and RGS16 promoters as promising candidates to support the delivery of cytokines such as IL-12 and IL-2 directly to the tumour site, leading to enhanced antitumour efficacy and long-term survival of mice in both syngeneic and xenogeneic models. This effect was concomitant with improved CAR T cell polyfunctionality, activation of endogenous antitumour immunity and a favourable safety profile, and was applicable in CAR T cells from patients.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: P.A.B. declares research funding from Bristol-Myers-Squibb. P.K.D. declares research funding from Myeloid Therapeutics, Prescient Therapeutics, Bristol-Myers-Squibb and Juno Therapeutics. I.A.P. declares research funding from AstraZeneca, Bristol-Myers-Squibb and Roche Genentech. The authors declare the following patents related to this work: PCT/AU2021/051219, ‘Composition and methods for immunotherapy’ (19 October 2021) (A.X.Y.C., I.G.H., P.K.D. and P.A.B.); and PCT/AU2024/050379, ‘Compositions & methods for immunotherapy – II’ (20 April 2023) (A.X.Y.C., K.M.Y., P.K.D. and P.A.B). M.H.P. serves on the Scientific Advisory Board of Allogene Therapeutics and Biogen. He is on the Board of Directors and has equity in Kamau Therapeutics and has equity in CRISPR Therapeutics. The remaining authors declare no competing interests.
Figures













References
-
- Albelda, S. M. CAR T cell therapy for patients with solid tumours: key lessons to learn and unlearn. Nat. Rev. Clin. Oncol.10.1038/s41571-023-00832-4 (2023). - PubMed
-
- Majzner, R. G. & Mackall, C. L. Clinical lessons learned from the first leg of the CAR T cell journey. Nat. Med.25, 1341–1355 (2019). - PubMed
-
- Mardiana, S., Solomon, B. J., Darcy, P. K. & Beavis, P. A. Supercharging adoptive T cell therapy to overcome solid tumor–induced immunosuppression. Sci. Transl. Med.11, eaaw2293 (2019). - PubMed
-
- Chmielewski, M. & Abken, H. TRUCKS, the fourth‐generation CAR T cells: current developments and clinical translation. Adv. Cell Gene Ther.10.1002/acg2.84 (2020).
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical

