Whole-genome ancestry of an Old Kingdom Egyptian
- PMID: 40604286
- PMCID: PMC12367555
- DOI: 10.1038/s41586-025-09195-5
Whole-genome ancestry of an Old Kingdom Egyptian
Abstract
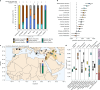
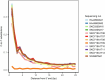
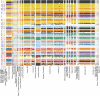
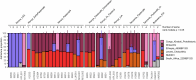
Ancient Egyptian society flourished for millennia, reaching its peak during the Dynastic Period (approximately 3150-30 BCE). However, owing to poor DNA preservation, questions about regional interconnectivity over time have not been addressed because whole-genome sequencing has not yet been possible. Here we sequenced a 2× coverage whole genome from an adult male Egyptian excavated at Nuwayrat (Nuerat, نويرات). Radiocarbon dated to 2855-2570 cal. BCE, he lived a few centuries after Egyptian unification, bridging the Early Dynastic and Old Kingdom periods. The body was interred in a ceramic pot within a rock-cut tomb1, potentially contributing to the DNA preservation. Most of his genome is best represented by North African Neolithic ancestry, among available sources at present. Yet approximately 20% of his genetic ancestry can be traced to genomes representing the eastern Fertile Crescent, including Mesopotamia and surrounding regions. This genetic affinity is similar to the ancestry appearing in Anatolia and the Levant during the Neolithic and Bronze Age2-5. Although more genomes are needed to fully understand the genomic diversity of early Egyptians, our results indicate that contacts between Egypt and the eastern Fertile Crescent were not limited to objects and imagery (such as domesticated animals and plants, as well as writing systems)6-9 but also encompassed human migration.
© 2025. The Author(s).
Conflict of interest statement
Competing interests: The authors declare no competing interests.
Figures







References
-
- Garstang, J. in The Burial Customs of Ancient Egypt (ed. Garstang, J.) 26–30 (Archibald Constable, 1907).
-
- Skourtanioti, E. et al. Genomic history of Neolithic to Bronze Age Anatolia, Northern Levant, and Southern Caucasus. Cell181, 1158–1175.e28 (2020). - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

