Gut microbiome compositional and functional features associate with Alzheimer's disease pathology
- PMID: 40604345
- PMCID: PMC12221809
- DOI: 10.1002/alz.70417
Gut microbiome compositional and functional features associate with Alzheimer's disease pathology
Abstract
Background: The gut microbiome is a potentially modifiable risk factor for Alzheimer's disease (AD); however, understanding of its composition and function regarding AD pathology is limited.
Methods: Shallow-shotgun metagenomics was used to analyze the fecal microbiome of participants in the Wisconsin Microbiome in Alzheimer's Risk Study, leveraging clinical data and cerebrospinal fluid (CSF) biomarkers. Differential abundance and ordinary least squares regression analyses were performed to find differentially abundant gut microbiome features and their associations with CSF biomarkers of AD and related pathologies.
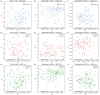
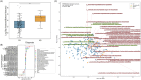
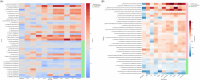
Results: Gut microbiome composition and function differed between individuals with and without AD dementia. The compositional difference was replicated in an independent cohort. Differentially abundant gut microbiome features were associated with CSF biomarkers of AD and related pathologies.
Discussion: These findings enhance our understanding of alterations in gut microbial composition and function in AD, and suggest that gut microbes and their pathways are linked to AD pathology.
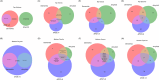
Highlights: Gut microbiome composition and function differ between people with Alzheimer's disease (AD) dementia and cognitively unimpaired (CU) individuals. Co-occurring gut microbes show differential abundance across AD-related groups (AD vs CU, amyloid status+ vs amyloid status-, and apolipoprotein E (APOE) ε4 status+ vs APOE ε4 status-). Gut microbiome composition also differs between people with AD dementia and CU individuals in a larger validation cohort. Differentially abundant gut microbiome composition and function between AD and CU groups are correlated with cerebrospinal fluid biomarkers for AD and related pathologies.
Keywords: Alzheimer's disease; biomarkers; cerebrospinal fluid; composition; differential abundance; function; gut microbiome; pathology.
© 2025 The Author(s). Alzheimer's & Dementia published by Wiley Periodicals LLC on behalf of Alzheimer's Association.
Conflict of interest statement
Dr. Kaddurah‐Daouk is an inventor of a series of patents on use of metabolomics for the diagnosis and treatment of central nervous system (CNS) diseases and holds equity in Metabolon Inc., Chymia LLC, and PsyProtix. Dr. Rob Knight is a scientific advisory board member and consultant for BiomeSense, Inc., and has equity and receives income. He is a scientific advisory board member and has equity in GenCirq. He is a consultant for DayTwo, and receives income. He has equity in and acts as a consultant for Cybele. He is a co‐founder of Biota, Inc., and has equity. He is a cofounder of Micronoma, and has equity and is a scientific advisory board member. The terms of these arrangements have been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies. Dr. Zetterberg has served on scientific advisory boards and/or as a consultant for Abbvie, Acumen, Alector, Alzinova, ALZPath, Amylyx, Annexon, Apellis, Artery Therapeutics, AZTherapies, Cognito Therapeutics, CogRx, Denali, Eisai, LabCorp, Merry Life, Nervgen, Novo Nordisk, Optoceutics, Passage Bio, Pinteon Therapeutics, Prothena, Red Abbey Labs, reMYND, Roche, Samumed, Siemens Healthineers, Triplet Therapeutics, and Wave; has given lectures in symposia sponsored by Alzecure, Biogen, Cellectricon, Fujirebio, Lilly, Novo Nordisk, and Roche; and is a co‐founder of Brain Biomarker Solutions in Gothenburg AB (BBS), which is a part of the GU Ventures Incubator Program (outside submitted work). Daniel McDonald is a consultant for, and has equity in, BiomeSence, Inc. The terms of this arrangement has been reviewed and approved by the University of California San Diego in accordance with its conflict of interest policies. All other authors have no conflicts of interest to disclose beyond those listed. Author disclosures are available in the Supporting Information.
Figures



Update of
-
Gut Microbiome Compositional and Functional Features Associate with Alzheimer's Disease Pathology.medRxiv [Preprint]. 2024 Sep 5:2024.09.04.24313004. doi: 10.1101/2024.09.04.24313004. medRxiv. 2024. Update in: Alzheimers Dement. 2025 Jul;21(7):e70417. doi: 10.1002/alz.70417. PMID: 39281749 Free PMC article. Updated. Preprint.
References
MeSH terms
Substances
Grants and funding
- #ADSF-21-831381-C/AD Strategic Fund and the Alzheimer's Association
- R01AG083883/AG/NIA NIH HHS/United States
- #101053962/European Union's Horizon Europe research and innovation programme
- #2022-01018/Swedish Research Council
- Olav Thon Foundation
- #2019-02397/Swedish Research Council
- U19 AG063744/AG/NIA NIH HHS/United States
- Bluefield Project
- #ADSF-21-831377-C/AD Strategic Fund and the Alzheimer's Association
- S10 OD026929/OD/NIH HHS/United States
- U19AG063744/AG/NIA NIH HHS/United States
- #ADSF-21-831376-C/AD Strategic Fund and the Alzheimer's Association
- #FO2022-0270/Familjen Rönströms Stiftelse, Stiftelsen för Gamla Tjänarinnor, Hjärnfonden, Sweden
- #22HLT07/European Partnership on Metrology co-financed from the European Union's Horizon Europe Research and Innovation Programme and by the Participating States
- #201809-2016862/Alzheimer Drug Discovery Foundation (ADDF)
- R01 AG070973/AG/NIA NIH HHS/United States
- JPND2021-00694/European Union Joint Programme-Neurodegenerative Disease Research
- #ADSF-24-1284328-C/AD Strategic Fund and the Alzheimer's Association
- Erling-Persson Family Foundation
- U01AG061359/AG/NIA NIH HHS/United States
- UKDRI-1003/NIH and Care Research University College London Hospitals Biomedical Research Centre and the UK Dementia Research Institute at UCL
- Vilas Early-Career Investigator Award
- #ALFGBG-71320/Swedish State Support for Clinical Research
- U01 AG061359/AG/NIA NIH HHS/United States
- Cure Alzheimer's Fund
- P30 AG062715/AG/NIA NIH HHS/United States
- #S10 OD026929/GF/NIH HHS/United States
- #2023-00356/Swedish Research Council
- R01AG070973/AG/NIA NIH HHS/United States
- #860197/European Union's Horizon 2020 research and innovation programme under the Marie Skłodowska-Curie
- R01 AG083883/AG/NIA NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

