H3K4 demethylase SsJMJ11 negatively regulates drought-tolerance responses in sugarcane
- PMID: 40604414
- PMCID: PMC12220489
- DOI: 10.1186/s12870-025-06832-z
H3K4 demethylase SsJMJ11 negatively regulates drought-tolerance responses in sugarcane
Abstract
Background: Drought-induced gene alteration is usually associated with changes of histone H3K4me3 in plants. Histone methylation homeostasis relies on the coordinated activity of methyltransferases and demethylases. We previously demonstrated that SsJMJ11 is an H3K4me3 demethylase in Saccharum spontaneum and participates in regulating flowering time. However, the role of H3K4me3 regulators in regulating drought-stress responses in sugarcane (Saccharum spp.) remains elusive.
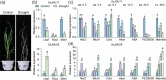
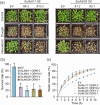
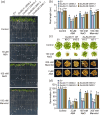
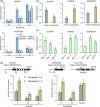
Results: We show that SsJMJ11 negatively regulates drought-stress responses by acting as an H3K4me3 demethylase. Ectopic overexpression of SsJMJ11 in Arabidopsis thaliana resulted in a hypersensitivity to soil drought stress as well as abscisic acid (ABA) and mannitol. Meanwhile, the drought-induced expression of AtRD20 and AtDREB2A, two well-known positive regulators of drought tolerance, was repressed by SsJMJ11 overexpression. In S. spontaneum, the ABA- and dehydration-induced transcription of SsRD20 and SsDREB2A was associated with increased H3K4me3 levels at these loci. Furthermore, transient overexpression of SsJMJ11 in S. spontaneum protoplasts reduced the ABA-induced transcription of SsRD20 and SsDREB2A, paralleling reduced H3K4me3 levels at these loci.
Conclusions: Our results suggest that SsJMJ11-mediated dynamic deposition of H3K4me3 is required for proper adaptation to drought stress in sugarcane.
Keywords: Saccharum spontaneum; Drought stress; H3K4me3; Histone demethylase; JmjC protein.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: Not applicable. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

Similar articles
-
H3K27 demethylase SsJMJ4 negatively regulates drought-stress responses in sugarcane.J Exp Bot. 2024 May 20;75(10):3040-3053. doi: 10.1093/jxb/erae037. J Exp Bot. 2024. PMID: 38310636
-
Modulation of an evolutionarily conserved epigenetic regulon controlling abscisic acid catabolism enhances drought tolerance in wheat.New Phytol. 2025 Aug;247(4):1777-1789. doi: 10.1111/nph.70302. Epub 2025 Jun 22. New Phytol. 2025. PMID: 40545778
-
Pepper JAZ Protein CaJAZ1-06 Negatively Regulates Drought Stress and Abscisic Acid Signaling.Physiol Plant. 2025 Jul-Aug;177(4):e70390. doi: 10.1111/ppl.70390. Physiol Plant. 2025. PMID: 40654046
-
Advancements in Water-Saving Strategies and Crop Adaptation to Drought: A Comprehensive Review.Physiol Plant. 2025 Jul-Aug;177(4):e70332. doi: 10.1111/ppl.70332. Physiol Plant. 2025. PMID: 40599019 Free PMC article. Review.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
References
-
- Zhang XT, Wang JG, Áin NY, Que YX, Zhang JS, Abid J, et al. International research initiative on genomics-guided sugarcane breeding. Mol Plant. 2025;S1674–2052(25):3–6.
-
- Lesk C, Rowhani P, Ramankutty N. Influence of extreme weather disasters on global crop production. Nature. 2016;529:84–7. - PubMed
-
- Zhang J, Zhang X, Tang H, Zhang Q, Hua X, Ma X, et al. Allele-defined genome of the autopolyploid sugarcane Saccharum spontaneum L. Nat Genet. 2018;50:1565–73. - PubMed
-
- Mall AK, Manimekalai R, Misra V, Pandey H, Srivastava S, Sharma A, et al. CRISPR/Cas-mediated genome editing for sugarcane improvement. Sugar Tech. 2024. 10.1007/s12355-023-01352-2.
-
- Mickelbart MV, Hasegawa PM, Bailey-Serres J. Genetic mechanisms of abiotic stress tolerance that translate to crop yield stability. Nat Rev Genet. 2015;16:237–51. - PubMed
MeSH terms
Substances
Grants and funding
- grant No. 32472070/National Natural Science Foundation of China
- grant No. 32472070/National Natural Science Foundation of China
- grant No. 32472070/National Natural Science Foundation of China
- grant No. 32472070/National Natural Science Foundation of China
- grant No. 32472070/National Natural Science Foundation of China
- grant No. 2022J01582/Natural Science Foundation of Fujian Province, China
- grant No. 2022J01582/Natural Science Foundation of Fujian Province, China
- grant No. 2022J01582/Natural Science Foundation of Fujian Province, China
- grant No. 2022J01582/Natural Science Foundation of Fujian Province, China
- grant No. 2022J01582/Natural Science Foundation of Fujian Province, China
- grant No. xjq202102/Distinguished Young Scholars of Fujian Agriculture and Forestry University
- grant No. xjq202102/Distinguished Young Scholars of Fujian Agriculture and Forestry University
- grant No. xjq202102/Distinguished Young Scholars of Fujian Agriculture and Forestry University
- grant No. xjq202102/Distinguished Young Scholars of Fujian Agriculture and Forestry University
- grant No. xjq202102/Distinguished Young Scholars of Fujian Agriculture and Forestry University
LinkOut - more resources
Full Text Sources
Miscellaneous

