Identification and functional characterization of JrbZIP40 in walnut reveals its role in salt and drought stress tolerance in transgenic Arabidopsis seedlings
- PMID: 40604448
- PMCID: PMC12218000
- DOI: 10.1186/s12870-025-06928-6
Identification and functional characterization of JrbZIP40 in walnut reveals its role in salt and drought stress tolerance in transgenic Arabidopsis seedlings
Abstract
Background: Salt and drought are the primary environmental stress factors that severely threaten plant growth, development, and yield. bZIP transcription factors reportedly play crucial roles in plant responses to both biotic and abiotic stressors. However, the biological function of bZIP transcription factors in oil crops, particularly walnuts, under salt and drought stress remains unclear.
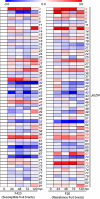
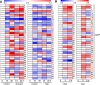
Results: In this study, members of the walnut bZIP gene family were identified based on the walnut genome Chandler 2.0. Transcriptome data and RT-qPCR results were used to analyze the expression patterns of various JrbZIP genes under biotic and abiotic stress, revealing that JrbZIP40 was strongly induced by both drought and salt stress. Subcellular localization and transcriptional activation assays demonstrated that JrbZIP40 localized to the nucleus and exhibited transcriptional activation activity. Overexpression of JrbZIP40 in transgenic Arabidopsis seedlings significantly enhanced resistance to salt and drought stress. DAP-seq and Dual-luciferase results indicated that JrbZIP40 may bind to the JrHB7 and JrATG8G promoters and activate their expression, contributing to stress resistance.
Conclusions: Overall, this study elucidates the regulatory network and biological functions of JrbZIP40 in drought and salt tolerance, providing a theoretical foundation and candidate gene resources for the future use of genetic engineering to improve walnut stress resistance.
Keywords: Juglans regia; Drought stress; Functional characterization; Salt stress; bZIP transcription factor.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: No specific permits were needed, and material collection and molecular experiments were carried out following current Chinese regulations. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures




Similar articles
-
Identification and expression of the AREB/ABF/ABI5 subfamily genes in chickpea and lentil reveal major players involved in ABA-mediated defense response to drought stress.Planta. 2025 Jun 10;262(1):22. doi: 10.1007/s00425-025-04740-y. Planta. 2025. PMID: 40493071
-
Genome-wide study and expression analysis of soybean ERF transcription factors and overexpression of GmERF205 enhances drought resistance in soybean.BMC Genomics. 2025 Aug 6;26(1):726. doi: 10.1186/s12864-025-11829-x. BMC Genomics. 2025. PMID: 40770289 Free PMC article.
-
Genome-wide characterization of GRAS gene family and their expression profiles under diverse biotic and abiotic stresses in Amorphophallus konjac.BMC Genomics. 2025 Jul 8;26(1):643. doi: 10.1186/s12864-025-11777-6. BMC Genomics. 2025. PMID: 40629278 Free PMC article.
-
NF-Y Transcription Factors: Key Players in Biotic and Abiotic Stress Tolerance in Plants.Bioessays. 2025 Aug;47(8):e70023. doi: 10.1002/bies.70023. Epub 2025 Jun 6. Bioessays. 2025. PMID: 40478603 Review.
-
A systematic review on the implications of concurrent heat and drought stress in modulating floral development in plants.Plant Sci. 2024 Dec;349:112248. doi: 10.1016/j.plantsci.2024.112248. Epub 2024 Sep 11. Plant Sci. 2024. PMID: 39265654
References
-
- Meshi T, Iwabuchi M. Plant transcription factors. Plant Cell Physiol. 1995;36(8):1405–20. 10.1093/oxfordjournals.pcp.a078903. - PubMed
-
- Ptashne M, Gann A. Transcriptional activation by recruitment. Nature. 1997;386:569–77. 10.1038/386569a0. - PubMed
-
- DrÖge-Laser W, Snoek BL, Snel B, Weiste C. The Arabidopsis bZIP transcription factor family-an update. Curr Opin Plant Biol. 2018;45:36–49. 10.1016/j.pbi.2018.05.001. - PubMed
-
- Jakoby M, Weisshaar B, DrÖge-Laser W, Vicente-Carbajosa J, Tiedemann J, Kroj T, et al. bZIP transcription factors in Arabidopsis. Trends Plant Sci. 2002;7(3):106–11. 10.1016/S1360-1385(01)02223-3. - PubMed
-
- Banerjee A, Roychoudhury A. Abscisic-acid-dependent basic leucine zipper (bZIP) transcription factors in plant abiotic stress. Protoplasma. 2017;254:3–16. 10.1007/s00709-015-0920-4. - PubMed
MeSH terms
Substances
Grants and funding
- 2025225/Hebei Agricultural University Student Innovation and Entrepreneurship Training Program Funding Project
- HBCT2025190205/Modern Agricultural Industry Technology System in Hebei Province-breeding and cultivation of novel walnut kinds
- 241790827A/Shijiazhuang Basic Research Project
- YJ2021050/Special Scientific Research Project for the Introduction of Talents in Hebei Agricultural University
LinkOut - more resources
Full Text Sources

