Oral microbiome diversity and composition before and after chemotherapy treatment in pediatric oncology patients
- PMID: 40604730
- PMCID: PMC12220142
- DOI: 10.1186/s12903-025-06405-4
Oral microbiome diversity and composition before and after chemotherapy treatment in pediatric oncology patients
Abstract
Objective: This study investigated the impact of anticancer treatment on the oral microbiome in pediatric patients and its association with oral mucositis (OM).
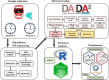
Materials and methods: A double-blind, randomized trial involving 34 pediatric cancer patients (ages 2-17.99) with solid or hematological malignancies. Mucosal swab samples were collected before and after chemotherapy. Patients underwent two 7-day rinse cycles-one with Caphosol and one with saline-in a randomized order. Bacterial DNA from 110 mucosal swabs was analyzed using 16S rRNA sequencing.
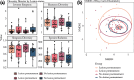
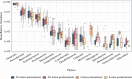
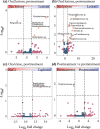
Results: Chemotherapy altered bacterial composition. No life-threatening OM cases (WHO grade 4) were observed, but mild to severe OM (grades 1-3) occurred in three patients. In patients without oral lesions, Bergeyella genus was more abundant prior to treatment while Alloprevotella was more abundant in the post-treatment samples, compared to patients with lesions. OM was linked to distinct microbiome profiles, including Stenotrophomonas, Leptotrichia sp., Serratia sp.,Capnocytophaga sputigena, Sphingomonas sp., Parapusillimonas sp., Staphylococcus sp., and Turicibacter genera. Additionally, Burkholderia-Caballeronia-Paraburkholderia (p = 0.013) were more prevalent in the Caphosol group compared to the saline group.
Conclusions: These findings indicate that chemotherapy-induced microbiome shifts associate with OM risk, highlighting the potential for microbial markers to predict high-risk patients and support protective strategies.
Trial registration: The trial titled "Supersaturated Calcium Phosphate Oral Rinse (Caphosol®) for the Prevention of Oral Mucositis in Children Undergoing Chemotherapeutic Treatments" was registered on ClinicalTrials.gov (ID NCT02807337), with the first submission date 2016-06-07.
Keywords: Chemotherapy; Diversity; Mucositis; Oral microbiome; Pediatric; Randomized.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: The study was approved by the Regional Ethics Committee of Tampere University Hospital in accordance with the principles of the Declaration of Helsinki. All patients and/or their parents provided written informed consent prior to participation. Every subject was given a study code number, and analyses were carried out without personal identification data. Consent for publication: Not applicable. Competing interests: The authors declare no competing interests.
Figures

References
Publication types
MeSH terms
Substances
Associated data
LinkOut - more resources
Full Text Sources
Medical

