An orally bioavailable BRD4 inhibitor disrupts expansion of a pathogenic epithelial-mesenchymal niche in bleomycin-induced fibrosis
- PMID: 40604997
- PMCID: PMC12225180
- DOI: 10.1186/s12931-025-03306-6
An orally bioavailable BRD4 inhibitor disrupts expansion of a pathogenic epithelial-mesenchymal niche in bleomycin-induced fibrosis
Abstract
Background: Idiopathic pulmonary fibrosis (IPF) is a relentlessly progressive and fatal disease with few effective therapies available. Fibrosis is driven, in part, by cell-state transitions of epithelial progenitors within the airways that repopulate the injured alveoli. This alveolar atypia affects gas exchange and stimulates ECM production. We sought to examine the role of BRD4 signaling in progenitor expansion in bleomycin-induced lung injury.
Methods: Activation of the Bromodomain-containing protein 4 (BRD4) epigenetic regulator in distinct stem cell populations was quantitated in a high-resolution scRNA-seq time course of bleomycin-induced injury, and confirmed in scRNA-seq studies in human IPF. A potent, selective, and orally bioavailable BRD4 inhibitor (BRD4i, ZL0969) was rationally designed and synthesized. The effect of BRD4i on myofibroblast transition, progenitor cell expansion and fibrosis was evaluated using a therapeutic experimental design in C57BL6/mice.
Results: We find that the BRD4 pathway is rapidly induced in regenerating activated alveolar type (AT)2 cells and persists in a population of pro-fibrotic Krt8 + progenitors expressing markers of epithelial mesenchymal transition as well as senescence. To test the functional role of BRD4 activation, we administered a potent, selective, and orally bioavailable BRD4 inhibitor (BRD4i, ZL0969) with ~ 80 nM IC50 to bleomycin-treated mice. BRD4i reduced myofibroblast formation and deposition of denatured ECM (collagen and laminin a1) in the alveolar space and improved disease scores. Importantly, BRD4i reduced a pathogenic population of alveolar progenitor cells expressing integrin (ITG)-A6/B4, tumor related protein 63 (Trp63) and keratin (Krt). In mice given an LD50 dose of bleomycin, BRD4 inhibition significantly improved their survival and reduced markers of disease.
Conclusions: These data demonstrate that inhibition of BRD4 signaling prevents expansion of myofibroblasts and expansion of a pathogenic epithelial progenitor population controlling alveolar atypia and fibrosis.
© 2025. The Author(s).
Conflict of interest statement
Declarations. Ethics approval and consent to participate: All vertebrate animals experiments were conducted using a protocol reviewed and approved by the University of Wisconsin-Madison School of Medicine and Public Health Institutional Animal Care and Use Committee (IACUC), adhering to guidelines set by National Institutes of Health (NIH) and Association for Assessment and Accreditation of Laboratory Animal Care (AAALAC). Consent for publication: Not applicable. Competing interests: JZ, BT and ARB hold patents on chemistry of BRD4 inhibitors. JZ and ARB are co-founders of Quadragenics Inc. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures




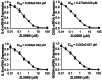
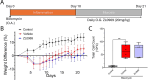






References
-
- Ley B, Collard HR, King TE. Clinical course and prediction of survival in idiopathic pulmonary fibrosis. Am J Respir Crit Care Med. 2011;183(4):431–40. - PubMed
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous

