Novel non-coding FOXP3 transcript isoform associated to potential transcriptional interference in human regulatory T cells
- PMID: 40605177
- PMCID: PMC12233831
- DOI: 10.1080/15476286.2025.2502719
Novel non-coding FOXP3 transcript isoform associated to potential transcriptional interference in human regulatory T cells
Abstract
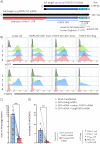
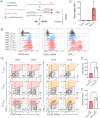
CD4+ regulatory T cells (TREGS) are critical for immune tolerance and the transcription factor Forkhead Box P3 (FOXP3) plays a crucial role in their differentiation and function. Recently, an alternative promoter has been reported for FOXP3, which is active only in TREGS and could have profound implications for the output of the locus, and therefore, for the functionality of these cells. By direct RNA sequencing we identified multiple novel FOXP3 transcriptional products, including one relatively abundant isoform with an extended 5' UTR that we named 'longFOXP3'. Western blotting, analysis of public mass spectrometry data, and transfection of in vitro transcribed RNA suggested that longFOXP3 is not coding. Furthermore, we show using two distinct RNA single-molecule fluorescence in situ hybridization technologies that transcription from the upstream promoter correlates with decreased levels of FOXP3 at the mRNA and protein levels. Together, we provide compelling evidence that the transcriptional output of the human FOXP3 locus is far more complex than that of the current annotation and warrants a more detailed analysis to identify coding and non-coding transcript isoforms. Furthermore, the alternative promoter may interfere with the activity of the canonical promoter, evoking intragenic transcriptional interference, and in this way, fine-tune the levels of FOXP3 in human TREGS.
Keywords: FOXP3; TREG; alternative promoter; transcript isoform; transcriptional interference.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
