Varying effects of chlorination on microbial functional repertoire and gene expression in contrasting effluents
- PMID: 40606158
- PMCID: PMC12213577
- DOI: 10.3389/fmicb.2025.1593147
Varying effects of chlorination on microbial functional repertoire and gene expression in contrasting effluents
Abstract
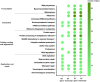
Effluents produced from different influent sources and sewage treatment processes carry distinct microbial community compositions. These microbiomes exhibit varying degrees of resistance and resilience under chlorination; however, their survival strategies and potential risks to the public health and ecosystem have yet to be fully characterized. In view of this, we subjected microbiomes from two contrasting types of effluents with distinct influent properties (seawater/freshwater-based) and prior treatment processes (primary/secondary) to metagenomics and metatranscriptomics analyses for comparing the alterations in their functional genes and activities under chlorination. The effluents presented highly dissimilar genomic and transcriptomic profiles. The variations in these profiles were significantly correlated to physicochemical factors including salinity, DO, BOD₅, TSS, and TN. We recovered novel metagenome-assembled genomes (MAGs) from each type of effluent, revealing that those recovered from the same effluent tended to share similar functional properties which aligned with the physicochemical parameters of the effluent. Notably, the type and extent of alterations in genomic and transcriptomic profiles under chlorination varied greatly between effluents. Most of the genes and transcripts with significant changes in relative abundances were exclusive to their respective effluents. Also, the number of genes and transcripts with significant increase in relative abundances after chlorination were much higher than those with reduction. These enriched genes and transcripts were responsible for a wide range of functions, including energy generation, repair of damaged components and stress responses. Furthermore, the remanent microbiomes in chlorinated effluents still harbored numerous genes related to waterborne diseases and antimicrobial resistance, suggesting the potential risks of discharging these effluents into the environment. This study revealed the diverse effects of chlorination on different types of effluent microbiomes. It suggested that the remanent microbiomes in chlorinated effluents would have great variance in genetic potential and activities, providing insights into the evaluation and regulation of chlorine disinfection in sewage treatment.
Keywords: chlorination; functional genes; gene expression; metagenomics; metatranscriptomics; microbiomes; sewage effluents.
Copyright © 2025 Tang and Lau.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest. The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Figures









References
-
- APHA (2005). Standard methods for the examination of water and wastewater. 21st Edn. Washington: American Public Health Association.
-
- Bensig E. O., Valadez-Cano C., Kuang Z. Y., Freire I. R., Reyes-Prieto A., MacLellan S. R. (2022). The two-component regulatory system CenK–CenR regulates expression of a previously uncharacterized protein required for salinity and oxidative stress tolerance in Sinorhizobium meliloti. Front. Microbiol. 13:1020932. doi: 10.3389/fmicb.2022.1020932, PMID: - DOI - PMC - PubMed
LinkOut - more resources
Full Text Sources

