Disruption of Lrpprc affects B cell development and proliferation in a mouse model of Leigh Syndrome French Canadian type
- PMID: 40607235
- PMCID: PMC12209026
- DOI: 10.1007/s44162-025-00094-x
Disruption of Lrpprc affects B cell development and proliferation in a mouse model of Leigh Syndrome French Canadian type
Abstract
Purpose: Leigh Syndrome French Canadian (LSFC) is a rare autosomal recessive metabolic disorder characterized by severe lactic acidosis crises and early mortality. LSFC patients carry variants in the Leucine Rich Pentatricopeptide Repeat Containing (LRPPRC) nuclear gene, which lead to defects in the respiratory chain complexes and mitochondrial dysfunction. Mitochondrial respiration modulates cellular metabolic activity, which impacts many cell processes, including the differentiation and function of immune cells. The purpose of this study is to define the role of Lrpprc on immune cell function.
Methods: As genetic deletion of Lrpprc is not viable, we generated two conditional mouse models: a model for systemic deletion of Lrpprc and a knock-in (KI) model carrying the most common LSFC pathogenic variant in Quebec, NM_133259.4(LRPPRC):c.1061C > T (p.Ala354Val).
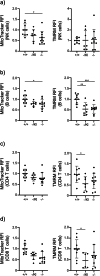
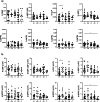
Results: We demonstrate that Lrpprc is an essential gene even in adult mice, as systemic deletion of Lrpprc leads to prominent weight loss and mortality. We also find an increase in lactate levels, a symptom of metabolic crises in LSFC. Lrpprc deletion and pathogenic variant affect various immune cell subsets, with a strong impact on B cell development and proliferation.
Conclusions: We generated a viable disease-relevant mouse model to study the role of Lrpprc in vivo and find that disruption of Lrpprc strongly impairs B cell development and proliferation.
Supplementary information: The online version contains supplementary material available at 10.1007/s44162-025-00094-x.
Keywords: B cells; Immune cells; LRPPRC; LSFC; Mitochondria; Proliferation.
© The Author(s) 2025.
Conflict of interest statement
Competing interestsThe authors declare no competing interests.
Figures







References
-
- Finsterer J. Leigh and Leigh-like syndrome in children and adults. Pediatr Neurol. 2008;39(4):223–35. - PubMed
-
- Debray FG, Morin C, Janvier A, Villeneuve J, Maranda B, Laframboise R, et al. LRPPRC mutations cause a phenotypically distinct form of Leigh syndrome with cytochrome c oxidase deficiency. J Med Genet. 2011;48(3):183–9. - PubMed
LinkOut - more resources
Full Text Sources
Research Materials
