Holoprosencephaly and cyclopia in bmp7b and bmpr1ba Crispant zebrafish
- PMID: 40607263
- PMCID: PMC12217118
- DOI: 10.1080/19768354.2025.2519018
Holoprosencephaly and cyclopia in bmp7b and bmpr1ba Crispant zebrafish
Abstract
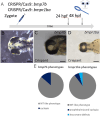
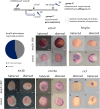
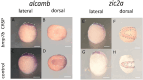
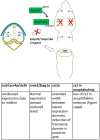
Holoprosencephaly (HPE) is the most frequent developmental disorder of the forebrain. In HPE, the early single anlage of the forebrain, the anterior neural plate (ANP) which encompasses the future telencephalon and eye field, fails to divide. BMP signaling and antagonism are overall important for nervous system development. The focus of this study was on the role of the ligand bmp7b and the receptor bmpr1ba during forebrain development. The zebrafish loci of bmp7b and bmpr1ba were targeted transiently with CRISPR/Ca9. Crispants for both bmp7b and bmpr1ba presented HPE and cyclopia, one central eye. Subsequently, the ANP was addressed in bmp7b Crispants. The morphology of the eye field was affected, with important markers, rx3, six3b and cxcr4a expressed condensed at the midline. Induced expression of bmp4 is also known to result in HPE. Such bmp4 induction altered the expression of bmpr1ba. Zebrafish Crispants for bmp7b and bmpr1ba can be used as a novel HPE model. A challenge in future analyses will be the penetrance of phenotypes in Crispants. The advantages are, however, that analyses can be conducted anywhere, without the need of mutant lines. One important aspect for future analysis will be the role of individual bmp ligands, receptors and antagonists in forebrain development.
Keywords: BMP; bmp7b; bmpr1ba; cyclopia; holoprosencephaly.
© 2025 The Author(s). Published by Informa UK Limited, trading as Taylor & Francis Group.
Conflict of interest statement
No potential conflict of interest was reported by the author(s).
Figures

References
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
