Precise Correction of the Pde6b-L659P Mutation Causing Retinal Degeneration with Minimum Bystander Editing by Advanced Genome Editing Tools
- PMID: 40607323
- PMCID: PMC12220932
- DOI: 10.34133/research.0770
Precise Correction of the Pde6b-L659P Mutation Causing Retinal Degeneration with Minimum Bystander Editing by Advanced Genome Editing Tools
Abstract
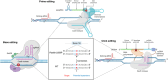
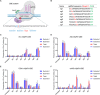
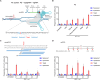
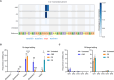
Recently developed base editing (BE), prime editing (PE), and click editing (CE) technologies enable precise and efficient genome editing with minimal risk of double-strand breaks and associated toxicity. However, their effectiveness in correcting real disease-causing mutations has not been systematically compared. Here, we aim to evaluate the potential of BE, PE, and CE technologies in rescuing the retinal degeneration-causing Pde6b (c.1976T>C, p.L659P) mutation. This site is prone to bystander effects, making it an ideal model for comparing the editing outcomes of these 3 novel technologies, particularly their editing precision. We optimized BE, PE, and CE systems in vitro using Pde6b-L659P cell models and compared their editing via deep sequencing. BE and PE had similar efficiency, but PE was the most precise, minimizing bystander edits. CE had lower efficiency and higher indel rates, needing further optimization. Using the optimal PE system for in vivo electroporation in Pde6b-L659P mice, we achieved 12.4% targeted repair with high precision, partially rescuing retinal degeneration. This study demonstrates proof of concept for the precise correction of the Pde6b-L659P mutation causing retinal degeneration using BE, PE, and CE tools. The findings offer valuable insights into the future optimization of precision gene editing techniques and their potential translational applications.
Copyright © 2025 Zhiquan Liu et al.
Conflict of interest statement
Competing interests: The authors declare that they have no competing interests.
Figures


Similar articles
-
Quality improvement strategies for diabetes care: Effects on outcomes for adults living with diabetes.Cochrane Database Syst Rev. 2023 May 31;5(5):CD014513. doi: 10.1002/14651858.CD014513. Cochrane Database Syst Rev. 2023. PMID: 37254718 Free PMC article.
-
Systemic pharmacological treatments for chronic plaque psoriasis: a network meta-analysis.Cochrane Database Syst Rev. 2021 Apr 19;4(4):CD011535. doi: 10.1002/14651858.CD011535.pub4. Cochrane Database Syst Rev. 2021. Update in: Cochrane Database Syst Rev. 2022 May 23;5:CD011535. doi: 10.1002/14651858.CD011535.pub5. PMID: 33871055 Free PMC article. Updated.
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of paclitaxel, docetaxel, gemcitabine and vinorelbine in non-small-cell lung cancer.Health Technol Assess. 2001;5(32):1-195. doi: 10.3310/hta5320. Health Technol Assess. 2001. PMID: 12065068
-
The clinical effectiveness and cost-effectiveness of enzyme replacement therapy for Gaucher's disease: a systematic review.Health Technol Assess. 2006 Jul;10(24):iii-iv, ix-136. doi: 10.3310/hta10240. Health Technol Assess. 2006. PMID: 16796930
-
A rapid and systematic review of the clinical effectiveness and cost-effectiveness of topotecan for ovarian cancer.Health Technol Assess. 2001;5(28):1-110. doi: 10.3310/hta5280. Health Technol Assess. 2001. PMID: 11701100
References
-
- Tsang SH, Sharma T. Retinitis pigmentosa (non-syndromic). Adv Exp Med Biol. 2018;1085:125–130. - PubMed
-
- Marconi S, Stout JT. PDE6B mutation-associated inherited retinal disease. Int Ophthalmol Clin. 2021;61(4):133–142. - PubMed
-
- Russell S, Bennett J, Wellman JA, Chung DC, Yu ZF, Tillman A, Wittes J, Pappas J, Elci O, McCague S, et al. Efficacy and safety of voretigene neparvovec (AAV2-hRPE65v2) in patients with RPE65-mediated inherited retinal dystrophy: A randomised, controlled, open-label, phase 3 trial. Lancet. 2017;390(10097):849–860. - PMC - PubMed
LinkOut - more resources
Full Text Sources

