ER stress genes (COL1A1, LOXL2, VWF) predicts IKK-16 as a Candidate therapeutic target for colitis-related inflammation and fibrosis suppression
- PMID: 40607383
- PMCID: PMC12213447
- DOI: 10.3389/fimmu.2025.1587860
ER stress genes (COL1A1, LOXL2, VWF) predicts IKK-16 as a Candidate therapeutic target for colitis-related inflammation and fibrosis suppression
Abstract
Introduction: The role of endoplasmic reticulum stress (ERS) in the immune-inflammatory dysregulation and intestinal fibrosis associated with ulcerative colitis (UC) remains unclear. This study aims to identify ERS-related genes involved in UC fibrosis and explore potential therapeutic targets.
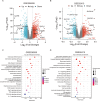
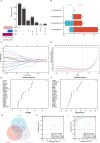
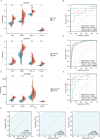
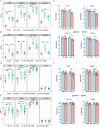
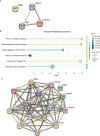
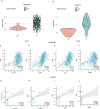
Methods: Differentially expressed ERS-related genes (DE-ERGs) were identified through comprehensive analysis of public datasets. Machine learning methods screened VWF, MZB1, COL1A1, and LOXL2 as key regulators. Immune infiltration analysis, protein-protein interaction (PPI) network analysis, and gene set variation analysis (GSVA) were performed to clarify their roles in UC fibrosis. Drug prediction was conducted using the Connectivity Map (CMap) database, supplemented by a literature review.
Results: The predicted drugs were ranked based on their binding affinities as follows: IKK-16 > Quercetin > Curcumin > Resveratrol > Budesonide > Trimebutine > Colchicine > Betamethasone > Pioglitazone > Metformin. IKK-16 showed the highest binding affinity for treating UC fibrosis. COL1A1, LOXL2, and VWF were identified as key drivers of UC intestinal fibrosis, supported by immune infiltration and PPI network analyses.
Discussion: These results suggest that ERS-related genes, particularly COL1A1, LOXL2, and VWF, may regulate UC fibrosis through interactions with immune cells. IKK-16 shows promise as a therapeutic agent. These findings provide new insights into UC pathogenesis and potential clinical treatment strategies.
Keywords: ER stress; bioinformatics analysis; immune infiltration; inflammation; intestinal fibrosis; molecular docking.
Copyright © 2025 Zhang, Yang, Yang, Guo, Huang, Cui, Wang, Wu and Wang.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Figures

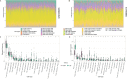




References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

