Structure of a new capsid form and comparison with A-, B-, and C-capsids clarify herpesvirus assembly
- PMID: 40607812
- PMCID: PMC12282113
- DOI: 10.1128/jvi.00504-25
Structure of a new capsid form and comparison with A-, B-, and C-capsids clarify herpesvirus assembly
Abstract
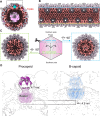
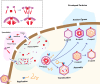
Three capsid types have been recognized from the nuclei of herpesvirus-infected cells: empty A-capsids, scaffolding-containing B-capsids, and DNA-filled C-capsids. Despite progress in determining atomic structures of these capsids and extracellular virions in recent years, debate persists concerning the origins and temporal relationships among these capsids during capsid assembly and genome packaging. Here, we have imaged over 300,000 capsids of herpes simplex virus type 1 by cryogenic electron microscopy (cryoEM) and exhaustively classified them to characterize the structural heterogeneity of the DNA-translocating portal complex and their functional states. The resultant atomic structures reveal not only the expected A-, B-, and C-capsids but also capsids with portal vertices similar to C-capsids but no resolvable genome in the capsid lumen, which we named D-capsids. The dodecameric dsDNA-translocating portal complex varies across these capsid types in their radial positions in icosahedral capsids and exhibits structural dynamics within each capsid type. In D-capsids, terminal DNA density exists in multiple conformations including one reminiscent of that in C-capsids, suggesting D-capsids are products of failed DNA retention. This interpretation is supported by varying amounts of DNA outside individual D-capsids and by the correlation of capsid counts observed in situ of infected cell nuclei and those after purification. Additionally, an "anchoring" segment of the scaffold protein is resolved interacting with the portal baskets of A- and B-capsids but not D- and C-capsids. Taken together, our data indicate that A-capsids arise from failed DNA packaging and D-capsids from failed genome retention, clarifying the origins of empty capsids in herpesvirus assembly.IMPORTANCEAs the prototypical herpesvirus, herpes simplex virus 1 (HSV-1) exhibits a global seroprevalence of 67% and approaching 90% in some localities. Herpesvirus infections can cause devastating cancers and birth defects, with HSV-1 infections leading to cold sores among the general population worldwide and blindness in developing nations. Here, we present atomic structures of the capsids sorted out from the nuclear isolates of HSV-1 infected cells, including the previously recognized A-, B-, and C-capsids, as well as the newly identified D-capsid. The structures show the details of protein-protein and protein-DNA interactions within each capsid type and the positional and interactional variability of the viral DNA-translocating portal vertex among these capsids. Importantly, our findings suggest that A-capsids are products of failed dsDNA packaging and D-capsids of failed genome retention. Together, the high-resolution 3D structures clarify the processes of genome packaging, maintenance, and ejection during capsid assembly, which are conserved across all herpesviruses.
Keywords: HSV-1; cryoEM; human herpesviruses; procapsid; virus assembly.
Conflict of interest statement
The authors declare no conflict of interest.
Figures




Update of
-
Structure of a new capsid form and comparison with A-, B- and C-capsids clarify herpesvirus assembly and DNA translocation.bioRxiv [Preprint]. 2025 May 8:2025.03.19.644230. doi: 10.1101/2025.03.19.644230. bioRxiv. 2025. Update in: J Virol. 2025 Jul 22;99(7):e0050425. doi: 10.1128/jvi.00504-25. PMID: 40166288 Free PMC article. Updated. Preprint.
Similar articles
-
Structure of a new capsid form and comparison with A-, B- and C-capsids clarify herpesvirus assembly and DNA translocation.bioRxiv [Preprint]. 2025 May 8:2025.03.19.644230. doi: 10.1101/2025.03.19.644230. bioRxiv. 2025. Update in: J Virol. 2025 Jul 22;99(7):e0050425. doi: 10.1128/jvi.00504-25. PMID: 40166288 Free PMC article. Updated. Preprint.
-
Internal Proteins of the Procapsid and Mature Capsids of Herpes Simplex Virus 1 Mapped by Bubblegram Imaging.J Virol. 2016 Apr 29;90(10):5176-86. doi: 10.1128/JVI.03224-15. Print 2016 May 15. J Virol. 2016. PMID: 26984725 Free PMC article.
-
The herpes simplex virus alkaline nuclease is required to maintain replication fork progression.J Virol. 2024 Dec 17;98(12):e0183624. doi: 10.1128/jvi.01836-24. Epub 2024 Nov 7. J Virol. 2024. PMID: 39508568 Free PMC article.
-
The Black Book of Psychotropic Dosing and Monitoring.Psychopharmacol Bull. 2024 Jul 8;54(3):8-59. Psychopharmacol Bull. 2024. PMID: 38993656 Free PMC article. Review.
-
Behavioral interventions to reduce risk for sexual transmission of HIV among men who have sex with men.Cochrane Database Syst Rev. 2008 Jul 16;(3):CD001230. doi: 10.1002/14651858.CD001230.pub2. Cochrane Database Syst Rev. 2008. PMID: 18646068
References
-
- McGeoch DJ. 1994. The human herpesviruses. Trends Microbiol 2:31–32. doi: 10.1016/0966-842X(94)90343-3 - DOI
MeSH terms
Substances
Grants and funding
- DBI-1338135/National Science Foundation
- T32 GM145388/GM/NIGMS NIH HHS/United States
- 5T32AI060567/NH/NIH HHS/United States
- S10RR23057/NH/NIH HHS/United States
- S10 OD018111/OD/NIH HHS/United States
- R01DE025567/NH/NIH HHS/United States
- R01 AI151055/AI/NIAID NIH HHS/United States
- U24 GM116792/GM/NIGMS NIH HHS/United States
- S10OD018111/NH/NIH HHS/United States
- T32 AR071307/AR/NIAMS NIH HHS/United States
- R01 DE025567/DE/NIDCR NIH HHS/United States
- T32 AI060567/AI/NIAID NIH HHS/United States
- S10 RR023057/RR/NCRR NIH HHS/United States
LinkOut - more resources
Full Text Sources

