Heterogeneous efflux pump expression underpins phenotypic resistance to antimicrobial peptides
- PMID: 40607907
- PMCID: PMC12226021
- DOI: 10.7554/eLife.99752
Heterogeneous efflux pump expression underpins phenotypic resistance to antimicrobial peptides
Abstract
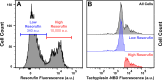
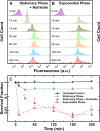
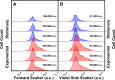
Antimicrobial resistance threatens the viability of modern medical interventions. There is a dire need to develop novel approaches to counter resistance mechanisms employed by starved or slow-growing pathogens that are refractory to conventional antimicrobial therapies. Antimicrobial peptides have been advocated as potential therapeutic solutions due to the low levels of genetic resistance observed in bacteria against these compounds. However, here we show that subpopulations of stationary phase Escherichia coli and Pseudomonas aeruginosa survive tachyplesin treatment without acquiring genetic mutations. These phenotypic variants display enhanced efflux activity to limit intracellular peptide accumulation. Differential regulation of genes involved in outer membrane vesicle secretion, membrane modification, and protease activity was also observed between phenotypically resistant and susceptible cells. We discovered that the formation of these phenotypic variants could be prevented by administering tachyplesin in combination with sertraline, a clinically used antidepressant, suggesting a novel approach for combatting antimicrobial-refractory stationary phase bacteria.
Keywords: E. coli; antibiotic accumulation; antibiotics; antimicrobial peptides; antimicrobial resistance; efflux pump inhibitors; efflux pumps; gene expression; heterogeneity; infectious disease; membrane transport; microbiology; microfluidics; physics of living systems; stationary phase; transcriptomics.
© 2024, Lee et al.
Conflict of interest statement
KL, UŁ, GT, MM, BZ, WP, AV, JW, AB, RY, PO, AF, AJ, Sv, PC, MB, BH, KT, SP No competing interests declared, BI is affiliated with PIPA LLC
Figures











Update of
- doi: 10.1101/2024.04.22.590445
- doi: 10.7554/eLife.99752.1
- doi: 10.7554/eLife.99752.2
- doi: 10.7554/eLife.99752.3
References
MeSH terms
Substances
Grants and funding
- BB/K003240/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- BB/V008021/1/BB_/Biotechnology and Biological Sciences Research Council/United Kingdom
- EP/T017856/1/Engineering and Physical Sciences Research Council
- MR/Y033892/1/MRC_/Medical Research Council/United Kingdom
- 10.35802/101650/WT_/Wellcome Trust/United Kingdom
LinkOut - more resources
Full Text Sources
Medical

