Hepcidin sustains Kupffer cell immune defense against bloodstream bacterial infection via gut-derived metabolites in mice
- PMID: 40607920
- PMCID: PMC12404757
- DOI: 10.1172/JCI189607
Hepcidin sustains Kupffer cell immune defense against bloodstream bacterial infection via gut-derived metabolites in mice
Abstract
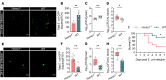
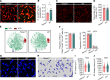
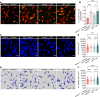
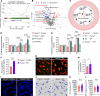
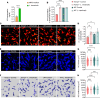
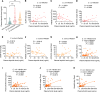
Bloodstream bacterial infections cause one-third of deaths from bacterial infections, and eradication of circulating bacteria is essential to prevent disseminated infections. Here, we found that hepcidin, the master regulator of systemic iron homeostasis, affected Kupffer cell (KC) immune defense against bloodstream bacterial infections by modulating the gut commensal bacteria-derived tryptophan derivative indole-3-propionic acid (IPA). Hepcidin deficiency impaired bacterial capture by KCs and exacerbated systemic bacterial dissemination through morphological changes in KCs. Gut microbiota depletion and fecal microbiota transplantation revealed that the gut microbiota mediated the alteration of KCs volume. Mechanistically, hepcidin deficiency led to a decreased abundance of the IPA-producing commensal Lactobacillus intestinalis and a concomitant reduction in the gut-to-liver shuttling of its metabolite IPA. IPA supplementation or L. intestinalis colonization restored the KC volume and hepatic immune defense against bloodstream bacterial infection in hepcidin-deficient mice. Moreover, hepcidin levels in patients with bacteremia were associated with days of antibiotic usage and hospitalization. Collectively, our findings highlight a previously unappreciated role of hepcidin in sustaining KC-mediated hepatic defense against bloodstream bacterial infections through the gut commensal L. intestinalis and its tryptophan derivative IPA. More importantly, we show that restoring the crosstalk between the gut microbiota and liver through IPA-inspired therapies may offer a promising strategy for enhancing the host defense against bloodstream bacterial infections in those with low hepcidin levels and a high risk for bacterial infections.
Keywords: Bacterial infections; Immunology; Infectious disease.
Figures


References
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

