Marburg virus glycoprotein mRNA vaccine is more protective than a virus-like particle-forming mRNA vaccine
- PMID: 40608418
- PMCID: PMC12490202
- DOI: 10.1172/JCI194586
Marburg virus glycoprotein mRNA vaccine is more protective than a virus-like particle-forming mRNA vaccine
Abstract
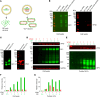
Although virus-like particle (VLP) vaccines were shown to be effective against several viruses, their advantage over vaccines that include envelope protein only is not completely clear, particularly for mRNA-encoded VLPs. We conducted a side-by-side comparison of the immunogenicity and protective efficacy of mRNA vaccines encoding the Marburg virus (MARV) full-length glycoprotein (GP) delivered alone or as a VLP. Electron microscopy confirmed VLP formation when MARV GP and matrix protein VP40 were coexpressed. We vaccinated guinea pigs with a 2-component mRNA vaccine encoding GP and VP40 (VLP) or GP alone. At the highest dose, both vaccines protected fully, although the VLP vaccine elicited a slightly lower humoral response than did the GP-only mRNA vaccine. However, at low doses, GP-only mRNA conferred 100% protection, whereas the VLP vaccine conferred only partial protection. In mice, VLP mRNA induced a moderate preference for GP-specific CD8+ T cell responses, whereas the GP-only mRNA somewhat favored CD4+ T cell responses. Guinea pig whole-blood RNA-Seq revealed that the VLP vaccine downregulated genes associated with various biological and metabolic processes, including the NF-κB signaling pathway, whereas the GP-only vaccine upregulated IFN signaling. Overall, the VLP mRNA vaccine was less immunogenic and protective, whereas the GP-only mRNA vaccine conferred robust protection with a dose of as little as 1 μg in guinea pigs.
Keywords: Cellular immune response; Immunoglobulins; Infectious disease; Vaccines; Virology.
Conflict of interest statement
Figures









References
-
- CDC. Marburg Outbreak in Rwanda Situation Summary. https://www.cdc.gov/marburg/situation-summary/index.html Updated December 2, 2024. Accessed July 10, 2025.
-
- WHO. Marburg virus disease – United Republic of Tanzania. https://www.who.int/emergencies/disease-outbreak-news/item/2025-DON559 Updated March 13, 2025. Accessed July 10, 2025.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Research Materials

