MICBG406A polymorphism reduces risk of mechanical ventilation and death during viral acute lung injury
- PMID: 40608426
- PMCID: PMC12333951
- DOI: 10.1172/jci.insight.191951
MICBG406A polymorphism reduces risk of mechanical ventilation and death during viral acute lung injury
Abstract
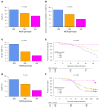
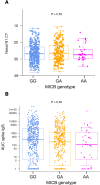
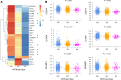
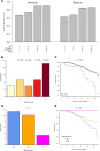
MHC class I polypeptide-related sequence B (MICB) is a ligand for NKG2D. We have shown NK cells are central to lung transplant acute lung injury (ALI) via NKG2D activation, and increased MICB in bronchoalveolar lavage predicts ALI severity. Separately, we found a MICB polymorphism (MICBG406A) is associated with decreased ALI risk. We hypothesized this polymorphism would protect against severe SARS-CoV-2 respiratory disease. We analyzed 1,036 patients hospitalized with SARS-CoV-2 infection from IMPACC. Associations between MICBG406A and outcomes were determined by linear regression or Cox proportional hazards models. We also measured immune profiles of peripheral blood and the upper and lower airway. We identified 560 major allele homozygous patients, and 426 and 50 with 1 or 2 copies of the variant allele, respectively. MICBG406A conferred reduced odds of severe COVID-19. MICBG406A homozygous participants demonstrated 34% reduced cumulative odds for mechanical ventilation or death and 43% reduced risk for mortality. Patients with MICBG406A variant alleles had reduced soluble inflammatory mediators and differential regulation of multiple immune pathways. These findings demonstrate a potentially novel association between increasing MICBG406A variant allele copies and reduced COVID-19 severity, independent of SARS-CoV-2 viral burden and humoral immunity, suggesting the NKG2D-ligand pathway as an intervention target.
Keywords: COVID-19; Immunology; Innate immunity; NK cells; Pulmonology.
Conflict of interest statement
Figures



References
MeSH terms
Substances
Grants and funding
- U19 AI090023/AI/NIAID NIH HHS/United States
- U01 AI167892/AI/NIAID NIH HHS/United States
- U19 AI057229/AI/NIAID NIH HHS/United States
- U19 AI062629/AI/NIAID NIH HHS/United States
- U19 AI128910/AI/NIAID NIH HHS/United States
- T32 DA018926/DA/NIDA NIH HHS/United States
- U19 AI125357/AI/NIAID NIH HHS/United States
- R01 AI145835/AI/NIAID NIH HHS/United States
- I01 BX005023/BX/BLRD VA/United States
- U19 AI128913/AI/NIAID NIH HHS/United States
- U19 AI118608/AI/NIAID NIH HHS/United States
- F32 HL165828/HL/NHLBI NIH HHS/United States
- U54 AI142766/AI/NIAID NIH HHS/United States
- R35 HL140026/HL/NHLBI NIH HHS/United States
- R01 AI122220/AI/NIAID NIH HHS/United States
- U19 AI077439/AI/NIAID NIH HHS/United States
- U19 AI118610/AI/NIAID NIH HHS/United States
- R01 AI068129/AI/NIAID NIH HHS/United States
- R01 AI135803/AI/NIAID NIH HHS/United States
- U19 AI089992/AI/NIAID NIH HHS/United States
- UM1 TR004528/TR/NCATS NIH HHS/United States
- IK2 BX005301/BX/BLRD VA/United States
- R01 AI104870/AI/NIAID NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

