Bioinformatics-guided construction of a tumor microenvironment-derived prognostic model in acute myeloid leukemia
- PMID: 40608798
- PMCID: PMC12225840
- DOI: 10.1371/journal.pone.0325145
Bioinformatics-guided construction of a tumor microenvironment-derived prognostic model in acute myeloid leukemia
Abstract
Background: The tumor microenvironment (TME) exerts a profound influence on the progression, therapeutic responses, and clinical outcomes of acute myeloid leukemia (AML), a prevalent hematologic malignancy in adults. This study aimed to establish a TME-based prognostic model to unveil novel therapeutic and prognostic avenues for AML.
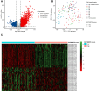
Methods: Gene expression profiles and clinical information for 134 AML patients were retrieved from The Cancer Genome Atlas (TCGA). The TME cellular components were evaluated using the ESTIMATE algorithm, and differentially expressed genes (DEGs) were identified. A Microenvironment Prognostic Model (MPM) was subsequently constructed through univariate Cox regression, LASSO regression, and multivariate Cox regression analyses. The predictive performance of the MPM was validated in a separate cohort of 312 AML patients from the TARGET database.
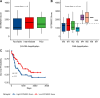
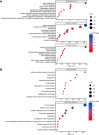
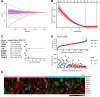
Results: Kaplan-Meier analysis revealed significant associations between the TME, French-American-British (FAB) classification, and overall survival (p-values = 3.6e-07 and 0.011, respectively). LASSO-Cox regression identified eight essential genes (CXCL12, GZMB, ITPR2, LYN, RAB9B, RGMB, RUFY4, TRIM16) that exhibited a strong correlation with survival (p-value < 0.0001). The MPM demonstrated excellent prognostic performance, with area under the curve (AUC) values of 84.05, 85.73, and 89.54 for predicting 1-, 3-, and 5-year survival, respectively. External validation with the TARGET database underscored the robustness of this model, yielding AUC values of 60.5%, 56.7%, and 55.7% at the corresponding intervals.
Conclusion: These findings present a TME-based prognostic model that offers a promising avenue for precise risk stratification and targeted therapeutic strategies in AML.
Copyright: © 2025 Navidinia et al. This is an open access article distributed under the terms of the Creative Commons Attribution License, which permits unrestricted use, distribution, and reproduction in any medium, provided the original author and source are credited.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures



References
-
- Bakhtiyari M, Liaghat M, Aziziyan F, Shapourian H, Yahyazadeh S, Alipour M, et al. The role of bone marrow microenvironment (BMM) cells in acute myeloid leukemia (AML) progression: immune checkpoints, metabolic checkpoints, and signaling pathways. Cell Commun Signal. 2023;21(1):252. doi: 10.1186/s12964-023-01282-2 - DOI - PMC - PubMed
-
- Cheung HL, Wong YH, Li YY, Yang X, Ko LH, Tan Kabigting JE, et al. Microenvironment matters: In vitro 3D bone marrow niches differentially modulate survival, phenotype and drug responses of acute myeloid leukemia (AML) cells. Biomaterials. 2025;312:122719. doi: 10.1016/j.biomaterials.2024.122719 - DOI - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Medical
Miscellaneous

